A new integrated framework for the identification of potential virus-drug associations
- PMID: 37675432
- PMCID: PMC10478006
- DOI: 10.3389/fmicb.2023.1179414
A new integrated framework for the identification of potential virus-drug associations
Abstract
Introduction: With the increasingly serious problem of antiviral drug resistance, drug repurposing offers a time-efficient and cost-effective way to find potential therapeutic agents for disease. Computational models have the ability to quickly predict potential reusable drug candidates to treat diseases.
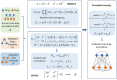
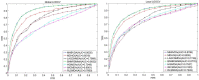
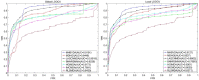
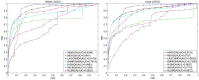
Methods: In this study, two matrix decomposition-based methods, i.e., Matrix Decomposition with Heterogeneous Graph Inference (MDHGI) and Bounded Nuclear Norm Regularization (BNNR), were integrated to predict anti-viral drugs. Moreover, global leave-one-out cross-validation (LOOCV), local LOOCV, and 5-fold cross-validation were implemented to evaluate the performance of the proposed model based on datasets of DrugVirus that consist of 933 known associations between 175 drugs and 95 viruses.
Results: The results showed that the area under the receiver operating characteristics curve (AUC) of global LOOCV and local LOOCV are 0.9035 and 0.8786, respectively. The average AUC and the standard deviation of the 5-fold cross-validation for DrugVirus datasets are 0.8856 ± 0.0032. We further implemented cross-validation based on MDAD and aBiofilm, respectively, to evaluate the performance of the model. In particle, MDAD (aBiofilm) dataset contains 2,470 (2,884) known associations between 1,373 (1,470) drugs and 173 (140) microbes. In addition, two types of case studies were carried out further to verify the effectiveness of the model based on the DrugVirus and MDAD datasets. The results of the case studies supported the effectiveness of MHBVDA in identifying potential virus-drug associations as well as predicting potential drugs for new microbes.
Keywords: Bounded Nuclear Norm Regularization; association prediction; drug; ensemble learning; matrix decomposition; virus.
Copyright © 2023 Qu, Song, Cheng, Jiang and Zhou.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
LinkOut - more resources
Full Text Sources

