ER-dependent membrane repair of mycobacteria-induced vacuole damage
- PMID: 37676004
- PMCID: PMC10653851
- DOI: 10.1128/mbio.00943-23
ER-dependent membrane repair of mycobacteria-induced vacuole damage
Abstract
Tuberculosis still remains a global burden and is one of the top infectious diseases from a single pathogen. Mycobacterium tuberculosis, the causative agent, has perfected many ways to replicate and persist within its host. While mycobacteria induce vacuole damage to evade the toxic environment and eventually escape into the cytosol, the host recruits repair machineries to restore the MCV membrane. However, how lipids are delivered for membrane repair is poorly understood. Using advanced fluorescence imaging and volumetric correlative approaches, we demonstrate that this involves the recruitment of the endoplasmic reticulum (ER)-Golgi lipid transfer protein OSBP8 in the Dictyostelium discoideum/Mycobacterium marinum system. Strikingly, depletion of OSBP8 affects lysosomal function accelerating mycobacterial growth. This indicates that an ER-dependent repair pathway constitutes a host defense mechanism against intracellular pathogens such as M. tuberculosis.
Keywords: Dictyostelium discoideum; Mycobacterium marinum; Mycobacterium tuberculosis; Sac1; lysosome; macrophages; membrane contact site; membrane repair; oxysterol-binding protein; phosphatidylinositol 4-phosphate; sterol.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
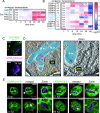
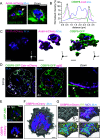
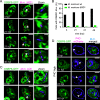
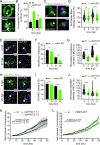

References
-
- Hanna N, Koliwer-Brandl H, Lefrançois LH, Kalinina V, Cardenal-Muñoz E, Appiah J, Leuba F, Gueho A, Hilbi H, Soldati T, Barisch C, Buchrieser C. 2021. Zn2+ intoxication of Mycobacterium marinum during Dictyostelium discoideum infection is counteracted by induction of the pathogen Zn2+ exporter CtpC. mBio 12:e01313-20. doi:10.1128/mBio.01313-20 - DOI - PMC - PubMed
-
- Stinear TP, Seemann T, Harrison PF, Jenkin GA, Davies JK, Johnson PDR, Abdellah Z, Arrowsmith C, Chillingworth T, Churcher C, Clarke K, Cronin A, Davis P, Goodhead I, Holroyd N, Jagels K, Lord A, Moule S, Mungall K, Norbertczak H, Quail MA, Rabbinowitsch E, Walker D, White B, Whitehead S, Small PLC, Brosch R, Ramakrishnan L, Fischbach MA, Parkhill J, Cole ST. 2008. Insights from the complete genome sequence of Mycobacterium marinum on the evolution of Mycobacterium tuberculosis. Genome Res 18:729–741. doi:10.1101/gr.075069.107 - DOI - PMC - PubMed
-
- Foulon M, Listian SA, Soldati T, Barisch C. 2022. Chapter 6 - conserved mechanisms drive host-lipid access, import, and utilization in Mycobacterium tuberculosis and M, p 133–161. In Fatima Z, Canaan S (ed), Biology of Mycobacterial lipids. Academic Press. doi:10.1016/B978-0-323-91948-7.00011-7 - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
