Coat protein of rice stripe virus enhances autophagy activity through interaction with cytosolic glyceraldehyde-3-phosphate dehydrogenases, a negative regulator of plant autophagy
- PMID: 37676568
- PMCID: PMC10441990
- DOI: 10.1007/s44154-023-00084-3
Coat protein of rice stripe virus enhances autophagy activity through interaction with cytosolic glyceraldehyde-3-phosphate dehydrogenases, a negative regulator of plant autophagy
Abstract
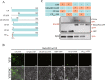
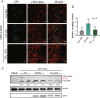
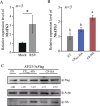
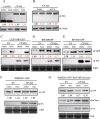
Viral infection commonly induces autophagy, leading to antiviral responses or conversely, promoting viral infection or replication. In this study, using the experimental plant Nicotiana benthamiana, we demonstrated that the rice stripe virus (RSV) coat protein (CP) enhanced autophagic activity through interaction with cytosolic glyceraldehyde-3-phosphate dehydrogenase 2 (GAPC2), a negative regulator of plant autophagy that binds to an autophagy key factor, autophagy-related protein 3 (ATG3). Competitive pull-down and co-immunoprecipitation (Co-IP)assays showed that RSV CP activated autophagy by disrupting the interaction between GAPC2 and ATG3. An RSV CP mutant that was unable to bind GAPC2 failed to disrupt the interaction between GAPC2 and ATG3 and therefore lost its ability to induce autophagy. RSV CP enhanced the autophagic degradation of a viral movement protein (MP) encoded by a heterologous virus, citrus leaf blotch virus (CLBV). However, the autophagic degradation of RSV-encoded MP and RNA-silencing suppressor (NS3) proteins was inhibited in the presence of CP, suggesting that RSV CP can protect MP and NS3 against autophagic degradation. Moreover, in the presence of MP, RSV CP could induce the autophagic degradation of a remorin protein (NbREM1), which negatively regulates RSV infection through the inhibition of viral cell-to-cell movement. Overall, our results suggest that RSV CP induces a selective autophagy to suppress the antiviral factors while protecting RSV-encoded viral proteins against autophagic degradation through an as-yet-unknown mechanism. This study showed that RSV CP plays dual roles in the autophagy-related interaction between plants and viruses.
Keywords: ATG3; Autophagy; Coat protein; GAPC; RSV.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no compete of interests.
Figures



Similar articles
-
Coat protein of Chinese wheat mosaic virus upregulates and interacts with cytosolic glyceraldehyde-3-phosphate dehydrogenase, a negative regulator of plant autophagy, to promote virus infection.J Integr Plant Biol. 2022 Aug;64(8):1631-1645. doi: 10.1111/jipb.13313. J Integr Plant Biol. 2022. PMID: 35713231
-
Rice Stripe Virus Interferes with S-acylation of Remorin and Induces Its Autophagic Degradation to Facilitate Virus Infection.Mol Plant. 2018 Feb 5;11(2):269-287. doi: 10.1016/j.molp.2017.11.011. Epub 2017 Dec 9. Mol Plant. 2018. PMID: 29229567
-
The plant protein NbP3IP directs degradation of Rice stripe virus p3 silencing suppressor protein to limit virus infection through interaction with the autophagy-related protein NbATG8.New Phytol. 2021 Jan;229(2):1036-1051. doi: 10.1111/nph.16917. Epub 2020 Oct 4. New Phytol. 2021. PMID: 32898938
-
Cotton leaf curl Multan virus βC1 Protein Induces Autophagy by Disrupting the Interaction of Autophagy-Related Protein 3 with Glyceraldehyde-3-Phosphate Dehydrogenases.Plant Cell. 2020 Apr;32(4):1124-1135. doi: 10.1105/tpc.19.00759. Epub 2020 Feb 12. Plant Cell. 2020. PMID: 32051213 Free PMC article.
-
Viruses and autophagy: bend, but don't break.Nat Rev Microbiol. 2024 May;22(5):309-321. doi: 10.1038/s41579-023-00995-y. Epub 2023 Dec 15. Nat Rev Microbiol. 2024. PMID: 38102460 Review.
Cited by
-
Autophagy plays an antiviral defence role against tomato spotted wilt orthotospovirus and is counteracted by viral effector NSs.Mol Plant Pathol. 2024 Oct;25(10):e70012. doi: 10.1111/mpp.70012. Mol Plant Pathol. 2024. PMID: 39350560 Free PMC article.
-
Beyond glycolysis: multifunctional roles of glyceraldehyde-3-phosphate dehydrogenases in plants.Hortic Res. 2025 Mar 3;12(6):uhaf070. doi: 10.1093/hr/uhaf070. eCollection 2025 Jun. Hortic Res. 2025. PMID: 40303431 Free PMC article.
-
Citrus tristeza virus p20 suppresses antiviral RNA silencing by co-opting autophagy-related protein 8 to mediate the autophagic degradation of SGS3.PLoS Pathog. 2025 Feb 24;21(2):e1012960. doi: 10.1371/journal.ppat.1012960. eCollection 2025 Feb. PLoS Pathog. 2025. PMID: 39993018 Free PMC article.
-
The Hypersensitive Response to Plant Viruses.Viruses. 2023 Sep 26;15(10):2000. doi: 10.3390/v15102000. Viruses. 2023. PMID: 37896777 Free PMC article. Review.
-
Integrated ATAC-seq and RNA-seq data analysis identifies transcription factors related to rice stripe virus infection in Oryza sativa.Mol Plant Pathol. 2024 Mar;25(3):e13446. doi: 10.1111/mpp.13446. Mol Plant Pathol. 2024. PMID: 38502176 Free PMC article.
References
-
- Cerff R (1978) Glyceraldehyde-3-phosphate dehydrogenase (NADP) from Sinapis alba: Steady state kinetics. Phytochemistry 17(12):2061–2067. 10.1016/S0031-9422(00)89281-X
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

