Genetic diversity of Salmonella enterica isolated over 13 years from raw California almonds and from an almond orchard
- PMID: 37676871
- PMCID: PMC10484465
- DOI: 10.1371/journal.pone.0291109
Genetic diversity of Salmonella enterica isolated over 13 years from raw California almonds and from an almond orchard
Abstract
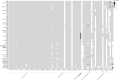
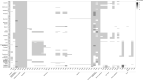
A comparative genomic analysis was conducted for 171 Salmonella isolates recovered from raw inshell almonds and raw almond kernels between 2001 and 2013 and for 30 Salmonella Enteritidis phage type (PT) 30 isolates recovered between 2001 and 2006 from a 2001 salmonellosis outbreak-associated almond orchard. Whole genome sequencing was used to measure the genetic distance among isolates by single nucleotide polymorphism (SNP) analyses and to predict the presence of plasmid DNA and of antimicrobial resistance (AMR) and virulence genes. Isolates were classified by serovars with Parsnp, a fast core-genome multi aligner, before being analyzed with the CFSAN SNP Pipeline (U.S. Food and Drug Administration Center for Food Safety and Applied Nutrition). Genetically similar (≤18 SNPs) Salmonella isolates were identified among several serovars isolated years apart. Almond isolates of Salmonella Montevideo (2001 to 2013) and Salmonella Newport (2003 to 2010) differed by ≤9 SNPs. Salmonella Enteritidis PT 30 isolated between 2001 and 2013 from survey, orchard, outbreak, and clinical samples differed by ≤18 SNPs. One to seven plasmids were found in 106 (62%) of the Salmonella isolates. Of the 27 plasmid families that were identified, IncFII and IncFIB plasmids were the most predominant. AMR genes were identified in 16 (9%) of the survey isolates and were plasmid encoded in 11 of 16 cases; 12 isolates (7%) had putative resistance to at least one antibiotic in three or more drug classes. A total of 303 virulence genes were detected among the assembled genomes; a plasmid that harbored a combination of pef, rck, and spv virulence genes was identified in 23% of the isolates. These data provide evidence of long-term survival (years) of Salmonella in agricultural environments.
Copyright: © 2023 Moyne et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures







References
-
- CDC Centers for Disease Control and Prevention. Outbreak of Salmonella serotype Enteritidis infections associated with raw almonds—United States and Canada, 2003–2004. MMWR Morb Mortal Wkly Rep. 2004: 53: 484–487. Available: https://www.cdc.gov/mmwr/preview/mmwrhtml/mm5322a8.htm. Accessed 11 Nov 2019. - PubMed

