Lower Airway Dysbiosis Augments Lung Inflammatory Injury in Mild-to-Moderate Chronic Obstructive Pulmonary Disease
- PMID: 37677136
- PMCID: PMC10867925
- DOI: 10.1164/rccm.202210-1865OC
Lower Airway Dysbiosis Augments Lung Inflammatory Injury in Mild-to-Moderate Chronic Obstructive Pulmonary Disease
Abstract
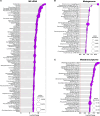
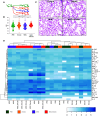
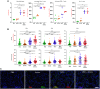
Rationale: Chronic obstructive pulmonary disease (COPD) is associated with high morbidity, mortality, and healthcare costs. Cigarette smoke is a causative factor; however, not all heavy smokers develop COPD. Microbial colonization and infections are contributing factors to disease progression in advanced stages. Objectives: We investigated whether lower airway dysbiosis occurs in mild-to-moderate COPD and analyzed possible mechanistic contributions to COPD pathogenesis. Methods: We recruited 57 patients with a >10 pack-year smoking history: 26 had physiological evidence of COPD, and 31 had normal lung function (smoker control subjects). Bronchoscopy sampled the upper airways, lower airways, and environmental background. Samples were analyzed by 16S rRNA gene sequencing, whole genome, RNA metatranscriptome, and host RNA transcriptome. A preclinical mouse model was used to evaluate the contributions of cigarette smoke and dysbiosis on lower airway inflammatory injury. Measurements and Main Results: Compared with smoker control subjects, microbiome analyses showed that the lower airways of subjects with COPD were enriched with common oral commensals. The lower airway host transcriptomics demonstrated differences in markers of inflammation and tumorigenesis, such as upregulation of IL-17, IL-6, ERK/MAPK, PI3K, MUC1, and MUC4 in mild-to-moderate COPD. Finally, in a preclinical murine model exposed to cigarette smoke, lower airway dysbiosis with common oral commensals augments the inflammatory injury, revealing transcriptomic signatures similar to those observed in human subjects with COPD. Conclusions: Lower airway dysbiosis in the setting of smoke exposure contributes to inflammatory injury early in COPD. Targeting the lower airway microbiome in combination with smoking cessation may be of potential therapeutic relevance.
Keywords: COPD; lung inflammation; metatranscriptomics; microbiome; transcriptomics.
Figures


Comment in
-
Investigating a Causal Role for Lung Microbiome Dysbiosis in Early Chronic Obstructive Pulmonary Disease Pathogenesis.Am J Respir Crit Care Med. 2023 Nov 15;208(10):1019-1021. doi: 10.1164/rccm.202309-1599ED. Am J Respir Crit Care Med. 2023. PMID: 37703423 Free PMC article. No abstract available.
References
-
- Sethi S, Murphy TF. Infection in the pathogenesis and course of chronic obstructive pulmonary disease. N Engl J Med . 2008;359:2355–2365. - PubMed
-
- Wilkinson TM, Patel IS, Wilks M, Donaldson GC, Wedzicha JA. Airway bacterial load and FEV1 decline in patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med . 2003;167:1090–1095. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- UL1 TR001445/TR/NCATS NIH HHS/United States
- P30 CA016087/CA/NCI NIH HHS/United States
- T32 ES007324/ES/NIEHS NIH HHS/United States
- U2C CA271890/CA/NCI NIH HHS/United States
- L30 AI138249/AI/NIAID NIH HHS/United States
- IK2 BX005309/BX/BLRD VA/United States
- K23 AI102970/AI/NIAID NIH HHS/United States
- R37 CA244775/CA/NCI NIH HHS/United States
- KL2 TR001446/TR/NCATS NIH HHS/United States
- R56 HL151700/HL/NHLBI NIH HHS/United States
- KL2TR001446/GF/NIH HHS/United States
- R21 GM147800/GM/NIGMS NIH HHS/United States
- R01 HL125816/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

