The sonication-assisted whisker method enables CRISPR-Cas9 ribonucleoprotein delivery to induce genome editing in rice
- PMID: 37679413
- PMCID: PMC10484913
- DOI: 10.1038/s41598-023-40433-w
The sonication-assisted whisker method enables CRISPR-Cas9 ribonucleoprotein delivery to induce genome editing in rice
Abstract
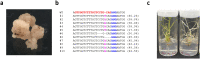
CRISPR/Cas9-based genome editing represents an unprecedented potential for plant breeding. Unlike animal cells, plant cells contain a rigid cell wall, genome editing tool delivery into plant cells is thus challenging. In particular, the delivery of the Cas9-gRNA ribonucleoprotein (RNP) into plant cells is desired since the transgene insertion into the genome should be avoided for industrial applications in plants. In this study, we present a novel RNP delivery approach in rice. We applied the sonication-assisted whisker method, conventionally developed for DNA delivery in plants, for RNP delivery in rice. Combined with marker gene delivery, we successfully isolated OsLCYβ genome-edited lines generated by RNPs. The calli and regenerated shoot of the OsLCYβ mutant showed abnormal carotenoid accumulation. In addition, we also detected, although at a low frequency, genome editing events in rice calli cells by RNP delivery using the sonication-assisted whisker method without any additional. Therefore, the sonication-assisted whisker method could be an attractive way to create RNP-based genome-edited lines in plants.
© 2023. Springer Nature Limited.
Conflict of interest statement
T.Y and T.T. have financial interests in Inplanta Innovations Inc. Yokohama, Japan. S.I. has financial interests in TOPPAN INC. A patent, which was invented by A.N., S.S.S., N.M., T.Y., T.T., and S.I., has been filed on behalf of JP2021-189460 by AIST, Inplanta Innovations Inc., and TOPPAN INC. M.F. has no conflict of interest.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

