CXCR6 orchestrates brain CD8+ T cell residency and limits mouse Alzheimer's disease pathology
- PMID: 37679549
- PMCID: PMC11102766
- DOI: 10.1038/s41590-023-01604-z
CXCR6 orchestrates brain CD8+ T cell residency and limits mouse Alzheimer's disease pathology
Abstract
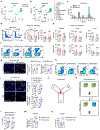
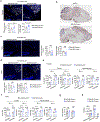
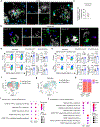
Neurodegenerative diseases, including Alzheimer's disease (AD), are characterized by innate immune-mediated inflammation, but functional and mechanistic effects of the adaptive immune system remain unclear. Here we identify brain-resident CD8+ T cells that coexpress CXCR6 and PD-1 and are in proximity to plaque-associated microglia in human and mouse AD brains. We also establish that CD8+ T cells restrict AD pathologies, including β-amyloid deposition and cognitive decline. Ligand-receptor interaction analysis identifies CXCL16-CXCR6 intercellular communication between microglia and CD8+ T cells. Further, Cxcr6 deficiency impairs accumulation, tissue residency programming and clonal expansion of brain PD-1+CD8+ T cells. Ablation of Cxcr6 or CD8+ T cells ultimately increases proinflammatory cytokine production from microglia, with CXCR6 orchestrating brain CD8+ T cell-microglia colocalization. Collectively, our study reveals protective roles for brain CD8+ T cells and CXCR6 in mouse AD pathogenesis and highlights that microenvironment-specific, intercellular communication orchestrates tissue homeostasis and protection from neuroinflammation.
© 2023. The Author(s), under exclusive licence to Springer Nature America, Inc.
Figures














Comment in
-
CD8+ T cells pump the brakes on Alzheimer's disease.Nat Immunol. 2023 Oct;24(10):1597-1598. doi: 10.1038/s41590-023-01622-x. Nat Immunol. 2023. PMID: 37679550 No abstract available.
References
Methods-only references
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

