Eukaryotic-like gephyrin and cognate membrane receptor coordinate corynebacterial cell division and polar elongation
- PMID: 37679597
- PMCID: PMC10522489
- DOI: 10.1038/s41564-023-01473-0
Eukaryotic-like gephyrin and cognate membrane receptor coordinate corynebacterial cell division and polar elongation
Abstract
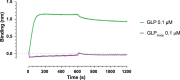
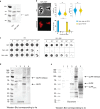
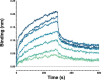
The order Corynebacteriales includes major industrial and pathogenic Actinobacteria such as Corynebacterium glutamicum or Mycobacterium tuberculosis. These bacteria have multi-layered cell walls composed of the mycolyl-arabinogalactan-peptidoglycan complex and a polar growth mode, thus requiring tight coordination between the septal divisome, organized around the tubulin-like protein FtsZ, and the polar elongasome, assembled around the coiled-coil protein Wag31. Here, using C. glutamicum, we report the discovery of two divisome members: a gephyrin-like repurposed molybdotransferase (Glp) and its membrane receptor (GlpR). Our results show how cell cycle progression requires interplay between Glp/GlpR, FtsZ and Wag31, showcasing a crucial crosstalk between the divisome and elongasome machineries that might be targeted for anti-mycobacterial drug discovery. Further, our work reveals that Corynebacteriales have evolved a protein scaffold to control cell division and morphogenesis, similar to the gephyrin/GlyR system that mediates synaptic signalling in higher eukaryotes through network organization of membrane receptors and the microtubule cytoskeleton.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






Update of
-
Eukaryotic-like gephyrin and cognate membrane receptor coordinate corynebacterial cell division and polar elongation.bioRxiv [Preprint]. 2023 Feb 1:2023.02.01.526586. doi: 10.1101/2023.02.01.526586. bioRxiv. 2023. Update in: Nat Microbiol. 2023 Oct;8(10):1896-1910. doi: 10.1038/s41564-023-01473-0. PMID: 36778425 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

