Unraveling virulence determinants in extended-spectrum beta-lactamase-producing Escherichia coli from East Africa using whole-genome sequencing
- PMID: 37679664
- PMCID: PMC10483776
- DOI: 10.1186/s12879-023-08579-0
Unraveling virulence determinants in extended-spectrum beta-lactamase-producing Escherichia coli from East Africa using whole-genome sequencing
Abstract
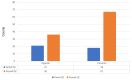
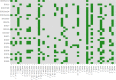
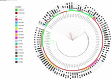
Escherichia coli significantly causes nosocomial infections and rampant spread of antimicrobial resistance (AMR). There is limited data on genomic characterization of extended-spectrum β-lactamase (ESBL)-producing E. coli from African clinical settings. This hospital-based longitudinal study unraveled the genetic resistance elements in ESBL E. coli isolates from Uganda and Tanzania using whole-genome sequencing (WGS). A total of 142 ESBL multi-drug resistant E. coli bacterial isolates from both Tanzania and Uganda were sequenced and out of these, 36/57 (63.1%) and 67/85 (78.8%) originated from Uganda and Tanzania respectively. Mutations in RarD, yaaA and ybgl conferring resistances to chloramphenicol, peroxidase and quinolones were observed from Ugandan and Tanzanian isolates. We reported very high frequencies for blaCTX-M-15 with 11/18(61.1%), and blaCTX-M-27 with 12/23 (52.1%), blaTEM-1B with 13/23 (56.5%) of isolates originating from Uganda and Tanzania respectively all conferring resistance to Beta-lactam-penicillin inhibitors. We observed chloramphenicol resistance-conferring gene mdfA in 21/23 (91.3%) of Tanzanian isolates. Extraintestinal E. coli sequence type (ST) 131 accounted for 5/59 (8.4%) of Tanzanian isolates while enterotoxigenic E. coli ST656 was reported in 9/34 (26.4%) of Ugandan isolates. Virulence factors originating from Shigella dysenteriae Sd197 (gspC, gspD, gspE, gspF, gspG, gspF, gspH, gspI), Yersinia pestis CO92 (irp1, ybtU, ybtX, iucA), Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 (csgF and csgG), and Pseudomonas aeruginosa PAO1 (flhA, fliG, fliM) were identified in these isolates. Overall, this study highlights a concerning prevalence and diversity of AMR-conferring elements shaping the genomic structure of multi-drug resistant E. coli in clinical settings in East Africa. It underscores the urgent need to strengthen infection-prevention controls and advocate for the routine use of WGS in national AMR surveillance and monitoring programs.Availability of WGS analysis pipeline: the rMAP source codes, installation, and implementation manual can free be accessed via https://github.com/GunzIvan28/rMAP .
Keywords: Antimicrobial resistance; East Africa; Extended-spectrum β-lactamase; Virulence factors; Whole-genome sequencing.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- Rossolini GM, Arena F, Pecile P. Pollini SJCoip. Update on the Antibiotic Resistance Crisis. 2014;18:56–60. - PubMed
-
- Nathan C, Cars OJNEJoM. Antibiotic resistance—problems, progress, and prospects. 2014;371(19):1761–3. - PubMed
-
- McEwen SA, Collignon PJ, Aarestrup FM, Schwarz S, Shen J, Cavaco L. Antimicrob Resistance: One Health Perspective. 2018;6(2):6. doi: 10.1128/microbiolspec.ARBA-0009-2017. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

