METTL3-mediated m6 A modification of circPRKAR1B promotes Crohn's colitis by inducing pyroptosis via autophagy inhibition
- PMID: 37679886
- PMCID: PMC10485333
- DOI: 10.1002/ctm2.1405
METTL3-mediated m6 A modification of circPRKAR1B promotes Crohn's colitis by inducing pyroptosis via autophagy inhibition
Abstract
Background: The roles of circRNA and N6-methyladenosine (m6 A) methylation in Crohn's disease (CD) have drawn much attention. Therefore, this investigation aimed to discover how the m6 A modification of circRNAs contributes to CD progression.
Methods: The study performed circRNA sequencing on colon samples from four CD patients and four normal controls (NCs) to screen for dysregulated circRNAs. Quantitative real-time polymerase chain reaction (qRT-PCR) was performed to validate the candidate circRNA expression and determine its correlation to CD-associated inflammatory indicators. In vivo and in vitro investigations were conducted to examine the functions and pathways of circPRKAR1B in CD, besides investigating the m6 A modification role in circRNA expression modulation.
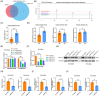
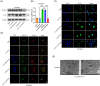
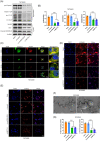
Results: The RNA-seq revealed that hsa_circ_0008039 (circPRKAR1B) was the most significant upregulated circRNA and was identified as the candidate circRNA for further examinations. Relative circPRKAR1B expression was significantly upregulated in CD colon tissues and closely related to CD-associated inflammatory indices. The circPRKAR1B expression and function were regulated by methyltransferase-like 3 (METTL3)-mediated m6 A methylation. In vitro studies indicated that circPRKAR1B promoted pyroptosis mediated by NLRP3 inflammasome (NLRP3; nucleotide-binding oligomerization domain, leucine-rich repeat and pyrin domain-containing 3) and impaired autophagy by interacting with the RNA-binding protein (RBP) SPTBN1, (SPTBN1; spectrin beta, non-erythrocytic 1). The in vivo investigations revealed the treatment effects of si-circPRKAR1B and si-METTL3 in colitis models of IL-10-deficient mice.
Conclusion: Our study reveals that METTL3-mediated m6 A modification of circPRKAR1B promotes Crohn's colitis by aggravating NLRP3 inflammasome-mediated pyroptosis via autophagy impairment in colonic epithelial cells.
Keywords: Crohn's disease; SPTBN1; autophagy; circPRKAR1B; m6A modification; pyroptosis.
© 2023 The Authors. Clinical and Translational Medicine published by John Wiley & Sons Australia, Ltd on behalf of Shanghai Institute of Clinical Bioinformatics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Roda G, Chien Ng S, Kotze PG, et al. Crohn's disease. Nat Rev Dis Primers. 2020;6:22. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
