A cross-institutional evaluation on breast cancer phenotyping NLP algorithms on electronic health records
- PMID: 37680211
- PMCID: PMC10480628
- DOI: 10.1016/j.csbj.2023.08.018
A cross-institutional evaluation on breast cancer phenotyping NLP algorithms on electronic health records
Abstract
Objective: Transformer-based language models are prevailing in the clinical domain due to their excellent performance on clinical NLP tasks. The generalizability of those models is usually ignored during the model development process. This study evaluated the generalizability of CancerBERT, a Transformer-based clinical NLP model, along with classic machine learning models, i.e., conditional random field (CRF), bi-directional long short-term memory CRF (BiLSTM-CRF), across different clinical institutes through a breast cancer phenotype extraction task.
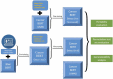
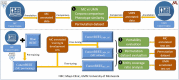
Materials and methods: Two clinical corpora of breast cancer patients were collected from the electronic health records from the University of Minnesota (UMN) and Mayo Clinic (MC), and annotated following the same guideline. We developed three types of NLP models (i.e., CRF, BiLSTM-CRF and CancerBERT) to extract cancer phenotypes from clinical texts. We evaluated the generalizability of models on different test sets with different learning strategies (model transfer vs locally trained). The entity coverage score was assessed with their association with the model performances.
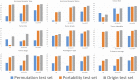
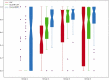
Results: We manually annotated 200 and 161 clinical documents at UMN and MC. The corpora of the two institutes were found to have higher similarity between the target entities than the overall corpora. The CancerBERT models obtained the best performances among the independent test sets from two clinical institutes and the permutation test set. The CancerBERT model developed in one institute and further fine-tuned in another institute achieved reasonable performance compared to the model developed on local data (micro-F1: 0.925 vs 0.932).
Conclusions: The results indicate the CancerBERT model has superior learning ability and generalizability among the three types of clinical NLP models for our named entity recognition task. It has the advantage to recognize complex entities, e.g., entities with different labels.
Keywords: Electronic health records; Generalizability; Information extraction; Natural language processing.
© 2023 The Authors. Published by Elsevier B.V. on behalf of Research Network of Computational and Structural Biotechnology.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
Similar articles
-
CancerBERT: a cancer domain-specific language model for extracting breast cancer phenotypes from electronic health records.J Am Med Inform Assoc. 2022 Jun 14;29(7):1208-1216. doi: 10.1093/jamia/ocac040. J Am Med Inform Assoc. 2022. PMID: 35333345 Free PMC article.
-
Extracting clinical named entity for pituitary adenomas from Chinese electronic medical records.BMC Med Inform Decis Mak. 2022 Mar 23;22(1):72. doi: 10.1186/s12911-022-01810-z. BMC Med Inform Decis Mak. 2022. PMID: 35321705 Free PMC article.
-
Extracting comprehensive clinical information for breast cancer using deep learning methods.Int J Med Inform. 2019 Dec;132:103985. doi: 10.1016/j.ijmedinf.2019.103985. Epub 2019 Oct 2. Int J Med Inform. 2019. PMID: 31627032
-
Named entity recognition from Chinese adverse drug event reports with lexical feature based BiLSTM-CRF and tri-training.J Biomed Inform. 2019 Aug;96:103252. doi: 10.1016/j.jbi.2019.103252. Epub 2019 Jul 16. J Biomed Inform. 2019. PMID: 31323311
-
Adversarial active learning for the identification of medical concepts and annotation inconsistency.J Biomed Inform. 2020 Aug;108:103481. doi: 10.1016/j.jbi.2020.103481. Epub 2020 Jul 18. J Biomed Inform. 2020. PMID: 32687985
Cited by
-
A taxonomy for advancing systematic error analysis in multi-site electronic health record-based clinical concept extraction.J Am Med Inform Assoc. 2024 Jun 20;31(7):1493-1502. doi: 10.1093/jamia/ocae101. J Am Med Inform Assoc. 2024. PMID: 38742455 Free PMC article.
-
Deep Learning Model for Natural Language to Assess Effectiveness of Patients With Non-Muscle Invasive Bladder Cancer Receiving Intravesical Bacillus Calmette-Guérin Therapy.JCO Clin Cancer Inform. 2025 Jun;9:e2400249. doi: 10.1200/CCI-24-00249. Epub 2025 Jun 27. JCO Clin Cancer Inform. 2025. PMID: 40577661 Free PMC article.
-
Clinical applications of large language models in medicine and surgery: A scoping review.J Int Med Res. 2025 Jul;53(7):3000605251347556. doi: 10.1177/03000605251347556. Epub 2025 Jul 4. J Int Med Res. 2025. PMID: 40615349 Free PMC article.
-
A text mining-based approach for comprehensive understanding of Chinese railway operational equipment failure reports.Sci Rep. 2025 Jul 30;15(1):27760. doi: 10.1038/s41598-025-11622-6. Sci Rep. 2025. PMID: 40739157 Free PMC article.
-
Multimodal deep learning for predicting neoadjuvant treatment outcomes in breast cancer: a systematic review.Biol Direct. 2025 Jun 23;20(1):72. doi: 10.1186/s13062-025-00661-8. Biol Direct. 2025. PMID: 40551237 Free PMC article.
References
-
- Carchiolo V., Longheu A., Reitano G., Zagarella L. 2019 Federated Conference on Computer Science and Information Systems (FedCSIS) IEEE,; 2019. Medical prescription classification: a NLP-based approach; pp. 605–609. Sep 1. Sep 1.
-
- Vijayakrishnan R., Steinhubl S.R., Ng K., Sun J., Byrd R.J., Daar Z., Williams B.A., Defilippi C., Ebadollahi S., Stewart W.F. Prevalence of heart failure signs and symptoms in a large primary care population identified through the use of text and data mining of the electronic health record. J Card Fail. 2014;20(7):459–464. Jul 1. - PMC - PubMed
-
- Mavrogiorgos K., Mavrogiorgou A., Kiourtis A., Zafeiropoulos N., Kleftakis S., Kyriazis D. 2022 32nd Conference of Open Innovations Association (FRUCT) IEEE,; 2022. Automated rule-based data cleaning using NLP; pp. 162–168. Nov 9. Nov 9.
Grants and funding
LinkOut - more resources
Full Text Sources
