Quantifying the arms race between LINE-1 and KRAB-zinc finger genes through TECookbook
- PMID: 37680368
- PMCID: PMC10480687
- DOI: 10.1093/nargab/lqad078
Quantifying the arms race between LINE-1 and KRAB-zinc finger genes through TECookbook
Abstract
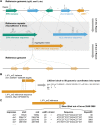
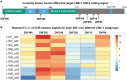
To defend against the invasion of transposons, hundreds of KRAB-zinc finger genes (ZNFs) evolved to recognize and silence various repeat families specifically. However, most repeat elements reside in the human genome with high copy numbers, making the ChIP-seq reads of ZNFs targeting these repeats predominantly multi-mapping reads. This complicates downstream data analysis and signal quantification. To better visualize and quantify the arms race between transposons and ZNFs, the R package TECookbook has been developed to lift ChIP-seq data into reference repeat coordinates with proper normalization and extract all putative ZNF binding sites from defined loci of reference repeats for downstream analysis. In conjunction with specificity profiles derived from in vitro Spec-seq data, human ZNF10 has been found to bind to a conserved ORF2 locus of selected LINE-1 subfamilies. This provides insight into how LINE-1 evaded capture at least twice and was subsequently recaptured by ZNF10 during evolutionary history. Through similar analyses, ZNF382 and ZNF248 were shown to be broad-spectrum LINE-1 binders. Overall, this work establishes a general analysis workflow to decipher the arms race between ZNFs and transposons through nucleotide substitutions rather than structural variations, particularly in the protein-coding region of transposons.
© The Author(s) 2023. Published by Oxford University Press on behalf of NAR Genomics and Bioinformatics.
Figures


References
-
- Imbeault M., Helleboid P.Y., Trono D. KRAB zinc-finger proteins contribute to the evolution of gene regulatory networks. Nature. 2017; 543:550–554. - PubMed
-
- de Tribolet-Hardy Jonas, Thorball ChristianW., Forey Romain, Planet Evarist, Duc Julien, Khubieh Bara, Offner Sandra, Fellay Jacques, Imbeault Michael, Turelli Priscillaet al.. Genetic features and genomic targets of human KRAB-Zinc Finger Proteins. 2023; bioRxiv doi:27 February 2023, preprint: not peer reviewed 10.1101/2023.02.27.530095. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

