The eukaryome of African children is influenced by geographic location, gut biogeography, and nutritional status
- PMID: 37680753
- PMCID: PMC10481997
- DOI: 10.1093/femsml/uqad033
The eukaryome of African children is influenced by geographic location, gut biogeography, and nutritional status
Abstract
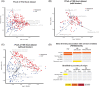
Eukaryotes have historically been studied as parasites, but recent evidence suggests they may be indicators of a healthy gut ecosystem. Here, we describe the eukaryome along the gastrointestinal tract of children aged 2-5 years and test for associations with clinical factors such as anaemia, intestinal inflammation, chronic undernutrition, and age. Children were enrolled from December 2016 to May 2018 in Bangui, Central African Republic and Antananarivo, Madagascar. We analyzed a total of 1104 samples representing 212 gastric, 187 duodenal, and 705 fecal samples using a metabarcoding approach targeting the full ITS2 region for fungi, and the V4 hypervariable region of the 18S rRNA gene for the overall eukaryome. Roughly, half of all fecal samples showed microeukaryotic reads. We find high intersubject variability, only a handful of taxa that are likely residents of the gastrointestinal tract, and frequent co-occurrence of eukaryotes within an individual. We also find that the eukaryome differs between the stomach, duodenum, and feces and is strongly influenced by country of origin. Our data show trends towards higher levels of Fusarium equiseti, a mycotoxin producing fungus, and lower levels of the protist Blastocystis in stunted children compared to nonstunted controls. Overall, the eukaryome is poorly correlated with clinical variables. Our study is of one of the largest cohorts analyzing the human intestinal eukaryome to date and the first to compare the eukaryome across different compartments of the gastrointestinal tract. Our results highlight the importance of studying populations across the world to uncover common features of the eukaryome in health.
Keywords: Sub-Saharan Africa; biogeography; fungi, protists; human intestinal eukaryome; microbiota.
© The Author(s) 2023. Published by Oxford University Press on behalf of FEMS.
Conflict of interest statement
None declared.
Figures




References
LinkOut - more resources
Full Text Sources
