A single-cell transcriptomic atlas characterizes age-related changes of murine cranial stem cell niches
- PMID: 37681346
- PMCID: PMC10652347
- DOI: 10.1111/acel.13980
A single-cell transcriptomic atlas characterizes age-related changes of murine cranial stem cell niches
Abstract
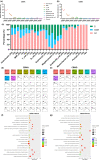
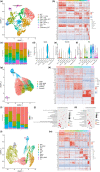
The craniofacial bones provide structural support for the skull and accommodate the vulnerable brain tissue with a protective cavity. The bone tissue undergoes constant turnover, which relies on skeletal stem cells (SSCs) and/or mesenchymal stem cells (MSCs) and their niches. SSCs/MSCs and their perivascular niche within the bone marrow are well characterized in long bones. As for cranial bones, besides bone marrow, the suture mesenchyme has been identified as a unique niche for SSCs/MSCs of craniofacial bones. However, a comprehensive study of the two different cranial stem cell niches at single-cell resolution is still lacking. In addition, during the progression of aging, age-associated changes in cranial stem cell niches and resident cells remain uncovered. In this study, we investigated age-related changes in cranial stem cell niches via single-cell RNA sequencing (scRNA-seq). The transcriptomic profiles and cellular compositions have been delineated, indicating alterations of the cranial bone marrow microenvironment influenced by inflammaging. Moreover, we identified a senescent mesenchymal cell subcluster and several age-related immune cell subclusters by reclustering and pseudotime trajectory analysis, which might be closely linked to inflammaging. Finally, differentially expressed genes (DEGs) and cell-cell communications were analyzed during aging, revealing potential regulatory factors. Overall, this work highlights the age-related changes in cranial stem cell niches, which deepens the current understanding of cranial bone and suture biology and may provide therapeutic targets for antiaging and regenerative medicine.
Keywords: cranial bone marrow; inflammaging; mesenchymal stem cell; single-cell RNA sequencing; stem cell niche; suture mesenchyme.
© 2023 The Authors. Aging Cell published by Anatomical Society and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Aldawood, Z. A. , Mancinelli, L. , Geng, X. , Yeh, S. A. , Di Carlo, R. , Leite, T. C. , Gustafson, J. , Wilk, K. , Yozgatian, J. , Garakani, S. , Bassir, S. H. , Cunningham, M. L. , Lin, C. P. , & Intini, G. (2023). Expansion of the sagittal suture induces proliferation of skeletal stem cells and sustains endogenous calvarial bone regeneration. Proceedings of the National Academy of Sciences, 120, e2120826120. - PMC - PubMed
-
- Almanan, M. , Raynor, J. , Ogunsulire, I. , Malyshkina, A. , Mukherjee, S. , Hummel, S. A. , Ingram, J. T. , Saini, A. , Xie, M. M. , Alenghat, T. , Way, S. S. , Deepe, G. S. , Divanovic, S. , Singh, H. , Miraldi, E. , Zajac, A. J. , Dent, A. L. , Hölscher, C. , Chougnet, C. , & Hildeman, D. A. (2020). IL‐10–producing Tfh cells accumulate with age and link inflammation with age‐related immune suppression. Science Advances, 6, eabb0806. - PMC - PubMed
-
- Ambrosi, T. H. , Marecic, O. , McArdle, A. , Sinha, R. , Gulati, G. S. , Tong, X. , Wang, Y. , Steininger, H. M. , Hoover, M. Y. , Koepke, L. S. , Murphy, M. P. , Sokol, J. , Seo, E. Y. , Tevlin, R. , Lopez, M. , Brewer, R. E. , Mascharak, S. , Lu, L. , Ajanaku, O. , … Chan, C. K. F. (2021). Aged skeletal stem cells generate an inflammatory degenerative niche. Nature, 597, 256–262. - PMC - PubMed
-
- Bronte, V. , Brandau, S. , Chen, S. H. , Colombo, M. P. , Frey, A. B. , Greten, T. F. , Mandruzzato, S. , Murray, P. J. , Ochoa, A. , Ostrand‐Rosenberg, S. , Rodriguez, P. C. , Sica, A. , Umansky, V. , Vonderheide, R. H. , & Gabrilovich, D. I. (2016). Recommendations for myeloid‐derived suppressor cell nomenclature and characterization standards. Nature Communications, 7, 12150. - PMC - PubMed
-
- Chan, C. K. F. , Gulati, G. S. , Sinha, R. , Tompkins, J. V. , Lopez, M. , Carter, A. C. , Ransom, R. C. , Reinisch, A. , Wearda, T. , Murphy, M. , Brewer, R. E. , Koepke, L. S. , Marecic, O. , Manjunath, A. , Seo, E. Y. , Leavitt, T. , Lu, W. J. , Nguyen, A. , Conley, S. D. , … Longaker, M. T. (2018). Identification of the human skeletal stem cell. Cell, 175, 43–56.e21. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

