The Highs and Lows of FBXW7: New Insights into Substrate Affinity in Disease and Development
- PMID: 37681873
- PMCID: PMC10486803
- DOI: 10.3390/cells12172141
The Highs and Lows of FBXW7: New Insights into Substrate Affinity in Disease and Development
Abstract
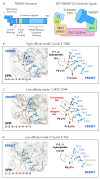
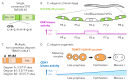
FBXW7 is a critical regulator of cell cycle, cell signaling, and development. A highly conserved F-box protein and component of the SKP1-Cullin-F-box (SCF) complex, FBXW7 functions as a recognition subunit within a Cullin-RING E3 ubiquitin ligase responsible for ubiquitinating substrate proteins and targeting them for proteasome-mediated degradation. In human cells, FBXW7 promotes degradation of a large number of substrate proteins, including many that impact disease, such as NOTCH1, Cyclin E, MYC, and BRAF. A central focus for investigation has been to understand the molecular mechanisms that allow the exquisite substrate specificity exhibited by FBXW7. Recent work has produced a clearer understanding of how FBXW7 physically interacts with both high-affinity and low-affinity substrates. We review new findings that provide insights into the consequences of "hotspot" missense mutations of FBXW7 that are found in human cancers. Finally, we discuss how the FBXW7-substrate interaction, and the kinases responsible for substrate phosphorylation, contribute to patterned protein degradation in C. elegans development.
Keywords: E3 ubiquitin ligase; FBXW7/FBW7/Cdc4/SEL-10; phospho-degron; ubiquitin–proteasome system.
Conflict of interest statement
The author declares no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

