Effects of Gestational Arsenic Exposures on Placental and Fetal Development in Mice: The Role of Cyr61
- PMID: 37682722
- PMCID: PMC10489955
- DOI: 10.1289/EHP12207
Effects of Gestational Arsenic Exposures on Placental and Fetal Development in Mice: The Role of Cyr61
Abstract
Background: Several epidemiological investigations demonstrated that maternal arsenic (As) exposure elevated risk of fetal growth restriction (FGR), but the mechanism remains unclear.
Objectives: This study aimed to investigate the effects of gestational As exposure on placental and fetal development and its underlying mechanism.
Methods: Dams were exposed to 0.15, 1.5, and throughout pregnancy via drinking water. Sizes of fetuses and placentas, placental histopathology, and glycogen content were measured. Placental RNA sequencing was conducted. Human trophoblasts were exposed to () to establish an in vitro model of As exposure. The mRNA stability and protein level of genes identified through RNA sequencing were measured. () modification was detected by methylated RNA immunoprecipitation-quantitative real-time polymerase chain reason (qPCR). The binding ability of insulin-like growth factor 2 binding protein 2 to the gene of interest was detected by RNA-binding protein immunoprecipitation-qPCR. Intracellular S-adenosylmethionine (SAM) and methyltransferase activity were determined by liquid chromatography-tandem mass spectrometry (LC-MS/MS) and colorimetry, respectively. In vitro methyltransferase (As3MT) knockdown or SAM supplementation and in vivo folic acid (FA) supplementation were used to evaluate the protective effect. A case-control study verified the findings.
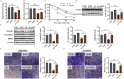
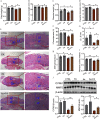
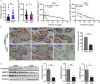
Results: Sizes of fetuses (exposed to 1.5 and ) and placentas (exposed to ) were lower in As-exposed mice. More trophoblasts accumulated and the expression of markers of interstitial invasion was lower in the mouse group in comparison with control. Placental RNA sequencing identified cysteine-rich angiogenic inducer 61 (Cyr61) as a candidate gene of interest. Mechanistically, mice and cells exposed to As had lower protein expression of CYR61, and this was attributed to a lower incidence of Cyr61 . Furthermore, cells exposed to As had lower methyltransferase activity, suggesting that this could be the mechanism by which Cyr61 was affected. Depletion of intracellular SAM, a cofactor for methyltransferase catalytic domain, partially contributed to As-induced methyltransferase activity reduction. Either As3MT knockdown or SAM supplementation attenuated As-induced Cyr61 down-regulation. In mice, FA supplementation rescued As-induced defective trophoblastic invasion and FGR. In humans, a negative correlation between maternal urinary As and plasma CYR61 was observed in infants who were small for gestational age.
Discussion: Using in vitro and in vivo models, we found that intracellular SAM depletion-mediated Cyr61 down-regulation partially contributed to As-induced defective trophoblastic invasion and FGR. https://doi.org/10.1289/EHP12207.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

