Immunocomplexed Antigen Capture and Identification by Native Top-Down Mass Spectrometry
- PMID: 37683262
- PMCID: PMC10557138
- DOI: 10.1021/jasms.3c00235
Immunocomplexed Antigen Capture and Identification by Native Top-Down Mass Spectrometry
Abstract
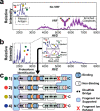
Antibody-antigen interactions are central to the immune response. Variation of protein antigens such as isoforms and post-translational modifications can alter their antibody binding sites. To directly connect the recognition of protein antigens with their molecular composition, we probed antibody-antigen complexes by using native tandem mass spectrometry. Specifically, we characterized the prominent peanut allergen Ara h 2 and a convergent IgE variable region discovered in patients who are allergic to peanuts. In addition to measuring the antigen-induced dimerization of IgE antibodies, we demonstrated how immunocomplexes can be isolated in the gas phase and activated to eject, identify, and characterize proteoforms of their bound antigens. Using tandem experiments, we isolated the ejected antigens and then fragmented them to identify their chemical composition. These results establish native top-down mass spectrometry as a viable platform for precise and thorough characterization of immunocomplexes to relate structure to function and enable the discovery of antigen proteoforms and their binding sites.
Keywords: Orbitrap; antibody; antigen; complex-up; native; top-down.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
-
- Skinner O. S.; Haverland N. A.; Fornelli L.; Melani R. D.; Do Vale L. H. F.; Seckler H. S.; Doubleday P. F.; Schachner L. F.; Srzentic K.; Kelleher N. L.; Compton P. D. Top-down characterization of endogenous protein complexes with native proteomics. Nat. Chem. Biol. 2018, 14, 36–41. 10.1038/nchembio.2515. - DOI - PMC - PubMed
-
- Anderson L. C.; English A. M.; Wang W.; Bai D. L.; Shabanowitz J.; Hunt D. F. Protein derivatization and sequential ion/ion reactions to enhance sequence coverage produced by electron transfer dissociation mass spectrometry. Int. J. Mass Spectrom. 2015, 377, 617–624. 10.1016/j.ijms.2014.06.023. - DOI - PMC - PubMed
-
- Haverland N. A.; Skinner O. S.; Fellers R. T.; Tariq A. A.; Early B. P.; LeDuc R. D.; Fornelli L.; Compton P. D.; Kelleher N. L. Defining Gas-Phase Fragmentation Propensities of Intact Proteins During Native Top-Down Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2017, 28, 1203–1215. 10.1007/s13361-017-1635-x. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

