eIF3d controls the persistent integrated stress response
- PMID: 37683648
- PMCID: PMC10528100
- DOI: 10.1016/j.molcel.2023.08.008
eIF3d controls the persistent integrated stress response
Abstract
Cells respond to intrinsic and extrinsic stresses by reducing global protein synthesis and activating gene programs necessary for survival. Here, we show that the integrated stress response (ISR) is driven by the non-canonical cap-binding protein eIF3d that acts as a critical effector to control core stress response orchestrators, the translation factor eIF2α and the transcription factor ATF4. We find that during persistent stress, eIF3d activates the translation of the kinase GCN2, inducing eIF2α phosphorylation and inhibiting general protein synthesis. In parallel, eIF3d upregulates the m6A demethylase ALKBH5 to drive 5' UTR-specific demethylation of stress response genes, including ATF4. Ultimately, this cascade converges on ATF4 expression by increasing mRNA engagement of translation machinery and enhancing ribosome bypass of upstream open reading frames (uORFs). Our results reveal that eIF3d acts in a life-or-death decision point during chronic stress and uncover a synergistic signaling mechanism in which translational cascades complement transcriptional amplification to control essential cellular processes.
Keywords: ATF4; GCN2; RNA methylation; eIF3d; integrated stress response; m(6)A; translation regulation.
Copyright © 2023 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures
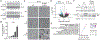



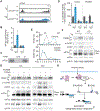
References
-
- Krishnamoorthy T, Pavitt GD, Zhang F, Dever TE, and Hinnebusch AG (2001). Tight binding of the phosphorylated alpha subunit of initiation factor 2 (eIF2ɑlpha) to the regulatory subunits of guanine nucleotide exchange factor eIF2B is required for inhibition of translation initiation. Molecular and cellular biology 21, 5018–5030. 10.1128/MCB.21.15.5018-5030.2001. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

