Serotonin signaling regulates actomyosin contractility during morphogenesis in evolutionarily divergent lineages
- PMID: 37684231
- PMCID: PMC10491668
- DOI: 10.1038/s41467-023-41178-w
Serotonin signaling regulates actomyosin contractility during morphogenesis in evolutionarily divergent lineages
Abstract
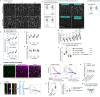
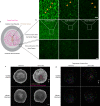
Serotonin is a neurotransmitter that signals through 5-HT receptors to control key functions in the nervous system. Serotonin receptors are also ubiquitously expressed in various organs and have been detected in embryos of different organisms. Potential morphogenetic functions of serotonin signaling have been proposed based on pharmacological studies but a mechanistic understanding is still lacking. Here, we uncover a role of serotonin signaling in axis extension of Drosophila embryos by regulating Myosin II (MyoII) activation, cell contractility and cell intercalation. We find that serotonin and serotonin receptors 5HT2A and 5HT2B form a signaling module that quantitatively regulates the amplitude of planar polarized MyoII contractility specified by Toll receptors and the GPCR Cirl. Remarkably, serotonin signaling also regulates actomyosin contractility at cell junctions, cellular flows and epiblast morphogenesis during chicken gastrulation. This phylogenetically conserved mechanical function of serotonin signaling in regulating actomyosin contractility and tissue flow reveals an ancestral role in morphogenesis of multicellular organisms.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Buznikov GA, Chudakova IV, Zvezdina ND. The r ole of neurohumours in early embryogenesis. i. serotonin content of developing embryos of sea urchin and loach. J. Embryol. Exp. Morphol. 1964;12:563–573. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

