Shared and distinct genetic etiologies for different types of clonal hematopoiesis
- PMID: 37684235
- PMCID: PMC10491829
- DOI: 10.1038/s41467-023-41315-5
Shared and distinct genetic etiologies for different types of clonal hematopoiesis
Abstract
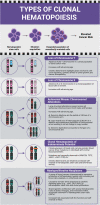
Clonal hematopoiesis (CH)-age-related expansion of mutated hematopoietic clones-can differ in frequency and cellular fitness by CH type (e.g., mutations in driver genes (CHIP), gains/losses and copy-neutral loss of chromosomal segments (mCAs), and loss of sex chromosomes). Co-occurring CH raises questions as to their origin, selection, and impact. We integrate sequence and genotype array data in up to 482,378 UK Biobank participants to demonstrate shared genetic architecture across CH types. Our analysis suggests a cellular evolutionary trade-off between different types of CH, with LOY occurring at lower rates in individuals carrying mutations in established CHIP genes. We observed co-occurrence of CHIP and mCAs with overlap at TET2, DNMT3A, and JAK2, in which CHIP precedes mCA acquisition. Furthermore, individuals carrying overlapping CH had high risk of future lymphoid and myeloid malignancies. Finally, we leverage shared genetic architecture of CH traits to identify 15 novel loci associated with leukemia risk.
© 2023. Springer Nature Limited.
Conflict of interest statement
V.G.S. serves as an advisor to and/or has equity in Branch Biosciences, Ensoma, Novartis, Forma, and Cellarity, all unrelated to the present work. All other authors declare no relevant competing interests.
Figures




References
-
- Mustjoki S, Young NS. Somatic mutations in ‘benign’ disease. N. Engl. J. Med. 2021;384:2039–2052. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

