A chromosome-level genome assembly of the darkbarbel catfish Pelteobagrus vachelli
- PMID: 37684295
- PMCID: PMC10491679
- DOI: 10.1038/s41597-023-02509-0
A chromosome-level genome assembly of the darkbarbel catfish Pelteobagrus vachelli
Abstract
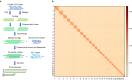
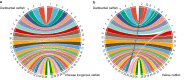
The darkbarbel catfish (Pelteobagrus vachelli), an economically important aquaculture species in China, is extensively employed in hybrid yellow catfish production due to its superior growth rate. However, information on its genome has been limited, constraining further genetic studies and breeding programs. Leveraging the power of PacBio long-read sequencing and Hi-C technologies, we present a high-quality, chromosome-level genome assembly for the darkbarbel catfish. The resulting assembly spans 692.10 Mb, with an impressive 99.9% distribution over 26 chromosomes. The contig N50 and scaffold N50 are 13.30 Mb and 27.55 Mb, respectively. The genome is predicted to contain 22,109 protein-coding genes, with 96.1% having functional annotations. Repeat elements account for approximately 35.79% of the genomic landscape. The completeness of darkbarbel catfish genome assembly is highlighted by a BUSCO score of 99.07%. This high-quality genome assembly provides a critical resource for future hybrid catfish breeding, comparative genomics, and evolutionary studies in catfish and other related species.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
A chromosome-level genome assembly of the male darkbarbel catfish (Pelteobagrus vachelli) using PacBio HiFi and Hi-C data.Sci Data. 2025 Feb 27;12(1):351. doi: 10.1038/s41597-025-04662-0. Sci Data. 2025. PMID: 40016230 Free PMC article.
-
Chromosomal-level assembly of yellow catfish genome using third-generation DNA sequencing and Hi-C analysis.Gigascience. 2018 Nov 1;7(11):giy120. doi: 10.1093/gigascience/giy120. Gigascience. 2018. PMID: 30256939 Free PMC article.
-
An improved chromosome-level genome assembly and annotation of Hong Kong catfish (Clarias fuscus).Sci Data. 2025 Feb 1;12(1):193. doi: 10.1038/s41597-025-04523-w. Sci Data. 2025. PMID: 39893196 Free PMC article.
-
Chromosome-Level Assembly of the Southern Rock Bream (Oplegnathus fasciatus) Genome Using PacBio and Hi-C Technologies.Front Genet. 2021 Dec 21;12:811798. doi: 10.3389/fgene.2021.811798. eCollection 2021. Front Genet. 2021. PMID: 34992639 Free PMC article. Review.
-
Retrospect and prospect of Nicotiana tabacum genome sequencing.Front Plant Sci. 2024 Sep 17;15:1474658. doi: 10.3389/fpls.2024.1474658. eCollection 2024. Front Plant Sci. 2024. PMID: 39354948 Free PMC article. Review.
Cited by
-
Low-Temperature Stress-Induced Hepatic Injury in Darkbarbel Catfish (Pelteobagrus vachelli): Mediated by Gut-Liver Axis Dysregulation.Antioxidants (Basel). 2025 Jun 21;14(7):762. doi: 10.3390/antiox14070762. Antioxidants (Basel). 2025. PMID: 40722866 Free PMC article.
-
A chromosome-level genome assembly of the male darkbarbel catfish (Pelteobagrus vachelli) using PacBio HiFi and Hi-C data.Sci Data. 2025 Feb 27;12(1):351. doi: 10.1038/s41597-025-04662-0. Sci Data. 2025. PMID: 40016230 Free PMC article.
-
Spatiotemporal Dynamics of Pro-Inflammatory Mediator Expression in Pelteobagrus vachelli During Ichthyophthiriasis: A 40-Day Longitudinal Study of IL-1β, IL-6, and SAA.Animals (Basel). 2025 May 28;15(11):1577. doi: 10.3390/ani15111577. Animals (Basel). 2025. PMID: 40509043 Free PMC article.
-
Chromosome-level genome assembly and annotation of the yellow grouper, Epinephelus awoara.Sci Data. 2024 Jan 31;11(1):151. doi: 10.1038/s41597-024-02989-8. Sci Data. 2024. PMID: 38296995 Free PMC article.
References
-
- Fricke, R., Eschmeyer, W. N. & Fong, J. D. Eschmeyer’s Catalog of Fishes: Genera/Species by Family/Subfamily. https://research.calacademy.org/research/ichthyology/catalog/SpeciesByFa.... Accessed 15 February 2023.
-
- Tacon AG. Trends in global aquaculture and aquafeed production: 2000–2017. Rev. Fish. Sci. Aquac. 2020;28:43–56. doi: 10.1080/23308249.2019.1649634. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

