Genomic epidemiology of SARS-CoV-2 from Uttar Pradesh, India
- PMID: 37684328
- PMCID: PMC10491582
- DOI: 10.1038/s41598-023-42065-6
Genomic epidemiology of SARS-CoV-2 from Uttar Pradesh, India
Erratum in
-
Author Correction: Genomic epidemiology of SARS-CoV-2 from Uttar Pradesh, India.Sci Rep. 2023 Oct 4;13(1):16723. doi: 10.1038/s41598-023-43636-3. Sci Rep. 2023. PMID: 37794066 Free PMC article. No abstract available.
Abstract
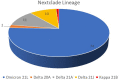
The various strains and mutations of SARS-CoV-2 have been tracked using several forms of genomic classification systems. The present study reports high-throughput sequencing and analysis of 99 SARS-CoV-2 specimens from Western Uttar Pradesh using sequences obtained from the GISAID database, followed by phylogeny and clade classification. Phylogenetic analysis revealed that Omicron lineages BA-2-like (55.55%) followed by Delta lineage-B.1.617.2 (45.5%) were predominantly circulating in this area Signature substitution at positions S: N501Y, S: D614G, S: T478K, S: K417N, S: E484A, S: P681H, and S: S477N were commonly detected in the Omicron variant-BA-2-like, however S: D614G, S: L452R, S: P681R and S: D950N were confined to Delta variant-B.1.617.2. We have also identified three escape variants in the S gene at codon position 19 (T19I/R), 484 (E484A/Q), and 681 (P681R/H) during the fourth and fifth waves in India. Based on the phylogenetic diversification studies and similar changes in other lineages, our analysis revealed indications of convergent evolution as the virus adjusts to the shifting immunological profile of its human host. To the best of our knowledge, this study is an approach to comprehensively map the circulating SARS-CoV-2 strains from Western Uttar Pradesh using an integrated approach of whole genome sequencing and phylogenetic analysis. These findings will be extremely valuable in developing a structured approach toward pandemic preparedness and evidence-based intervention plans in the future.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- Weekly update on COVID-19-21 August 2020. http://www.who.inthttps://www.who.int/publications/m/item/weekly-update-on-covid-19---16-o....
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

