SENP5 promotes homologous recombination-mediated DNA damage repair in colorectal cancer cells through H2AZ deSUMOylation
- PMID: 37684630
- PMCID: PMC10486113
- DOI: 10.1186/s13046-023-02789-9
SENP5 promotes homologous recombination-mediated DNA damage repair in colorectal cancer cells through H2AZ deSUMOylation
Abstract
Background: Neoadjuvant radiotherapy has been used as the standard treatment of colorectal cancer (CRC). However, radiotherapy resistance often results in treatment failure. To identify radioresistant genes will provide novel targets for combined treatments and prognostic markers.
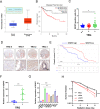
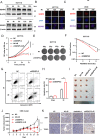
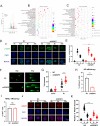
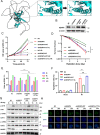
Methods: Through high content screening and tissue array from CRC patients who are resistant or sensitive to radiotherapy, we identified a potent resistant gene SUMO specific peptidase 5 (SENP5). Then, the effect of SENP5 on radiosensitivity was investigated by CCK8, clone formation, comet assay, immunofluorescence and flow cytometric analysis of apoptosis and cell cycle to investigate the effect of SENP5 on radiosensitivity. SUMO-proteomic mass spectrometry combined with co-immunoprecipitation assay were used to identify the targets of SENP5. Patient-derived organoids (PDO) and xenograft (PDX) models were used to explore the possibility of clinical application.
Results: We identified SENP5 as a potent radioresistant gene through high content screening and CRC patients tissue array analysis. Patients with high SENP5 expression showed increased resistance to radiotherapy. In vitro and in vivo experiments demonstrated that SENP5 knockdown significantly increased radiosensitivity in CRC cells. SENP5 was further demonstrated essential for efficient DNA damage repair in homologous recombination (HR) dependent manner. Through SUMO mass spectrometry analysis, we characterized H2AZ as a deSUMOylation substrate of SENP5, and depicted the SUMOylation balance of H2AZ in HR repair and cancer resistance. By using PDO and PDX models, we found targeting SENP5 significantly increased the therapeutic efficacy of radiotherapy.
Conclusion: Our findings revealed novel role of SENP5 in HR mediated DNA damage repair and cancer resistance, which could be applied as potent prognostic marker and intervention target for cancer radiotherapy.
Keywords: Cancer resistance; DNA damage repair; SENP5; deSUMOylation.
© 2023. Italian National Cancer Institute ‘Regina Elena’.
Conflict of interest statement
The authors declare no potential conflicts of interest.
Figures




References
MeSH terms
Grants and funding
- 82273579/National Natural Science Foundation of China
- 82173462/National Natural Science Foundation of China
- 82103776/National Natural Science Foundation of China
- 81972968/National Natural Science Foundation of China
- 82203979/National Natural Science Foundation of China
- GWV-10.2-YQ27/Three-Year Plan of Shanghai Municipal Health Commission
- No. 20214Y0012/three-year plan of shanghai municipal health commission
- 23QA1114600/Shanghai Science and Technology Development Foundation
- No.ZFY-2021-K-002/South Zhejiang Institute of Radiation Medicine and Nuclear Technology
- 2020AY30010/Science and Technology Project of Jiaxing City
- LQ23H220001/Zhejiang Provincial Natural Science Foundation of China
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

