Retro-miRs: novel and functional miRNAs originating from mRNA retrotransposition
- PMID: 37684690
- PMCID: PMC10486083
- DOI: 10.1186/s13100-023-00301-w
Retro-miRs: novel and functional miRNAs originating from mRNA retrotransposition
Abstract
Background: Reverse-transcribed gene copies (retrocopies) have emerged as major sources of evolutionary novelty. MicroRNAs (miRNAs) are small and highly conserved RNA molecules that serve as key post-transcriptional regulators of gene expression. The origin and subsequent evolution of miRNAs have been addressed but not fully elucidated.
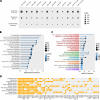
Results: In this study, we performed a comprehensive investigation of miRNA origination through retroduplicated mRNA sequences (retro-miRs). We identified 17 retro-miRs that emerged from the mRNA retrocopies. Four of these retro-miRs had de novo origins within retrocopied sequences, while 13 retro-miRNAs were located within exon regions and duplicated along with their host mRNAs. We found that retro-miRs were primate-specific, including five retro-miRs conserved among all primates and two human-specific retro-miRs. All retro-miRs were expressed, with predicted and experimentally validated target genes except miR-10527. Notably, the target genes of retro-miRs are involved in key biological processes such as metabolic processes, cell signaling, and regulation of neurotransmitters in the central nervous system. Additionally, we found that these retro-miRs play a potential oncogenic role in cancer by targeting key cancer genes and are overexpressed in several cancer types, including liver hepatocellular carcinoma and stomach adenocarcinoma.
Conclusions: Our findings demonstrated that mRNA retrotransposition is a key mechanism for the generation of novel miRNAs (retro-miRs) in primates. These retro-miRs are expressed, conserved, have target genes with important cellular functions, and play important roles in cancer.
Keywords: Cancer; Gene expression; Gene origination; Retrocopies; miRNAs.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Grants and funding
LinkOut - more resources
Full Text Sources

