Differential Type-I Interferon Response in Buffy Coat Transcriptome of Individuals Infected with SARS-CoV-2 Gamma and Delta Variants
- PMID: 37685953
- PMCID: PMC10487928
- DOI: 10.3390/ijms241713146
Differential Type-I Interferon Response in Buffy Coat Transcriptome of Individuals Infected with SARS-CoV-2 Gamma and Delta Variants
Abstract
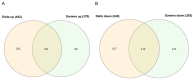
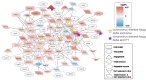
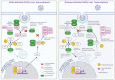
The innate immune system is the first line of defense against pathogens such as the acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The type I-interferon (IFN) response activation during the initial steps of infection is essential to prevent viral replication and tissue damage. SARS-CoV and SARS-CoV-2 can inhibit this activation, and individuals with a dysregulated IFN-I response are more likely to develop severe disease. Several mutations in different variants of SARS-CoV-2 have shown the potential to interfere with the immune system. Here, we evaluated the buffy coat transcriptome of individuals infected with Gamma or Delta variants of SARS-CoV-2. The Delta transcriptome presents more genes enriched in the innate immune response and Gamma in the adaptive immune response. Interactome and enriched promoter analysis showed that Delta could activate the INF-I response more effectively than Gamma. Two mutations in the N protein and one in the nsp6 protein found exclusively in Gamma have already been described as inhibitors of the interferon response pathway. This indicates that the Gamma variant evolved to evade the IFN-I response. Accordingly, in this work, we showed one of the mechanisms that variants of SARS-CoV-2 can use to avoid or interfere with the host Immune system.
Keywords: Delta; Gamma; SARS-CoV-2 variants; innate immune system; type-I interferon response; viral evolution.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

