Revealing the SARS-CoV-2 Spike Protein and Specific Antibody Immune Complex Formation Mechanism for Precise Evaluation of Antibody Affinity
- PMID: 37686023
- PMCID: PMC10487573
- DOI: 10.3390/ijms241713220
Revealing the SARS-CoV-2 Spike Protein and Specific Antibody Immune Complex Formation Mechanism for Precise Evaluation of Antibody Affinity
Abstract
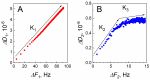
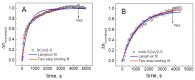
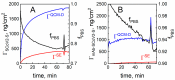
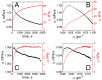
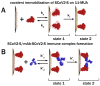
The profound understanding and detailed evaluation of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike (SCoV2-S) protein and specific antibody interaction mechanism is of high importance in the development of immunosensors for COVID-19. In the present work, we studied a model system of immobilized SCoV2-S protein and specific monoclonal antibodies by molecular dynamics of immune complex formation in real time. We simultaneously applied spectroscopic ellipsometry and quartz crystal microbalance with dissipation to reveal the features and steps of the immune complex formation. We showed direct experimental evidence based on acoustic and optical measurements that the immune complex between covalently immobilized SCoV2-S and specific monoclonal antibodies is formed in two stages. Based on these findings it was demonstrated that applying a two-step binding mathematical model for kinetics analysis leads to a more precise determination of interaction rate constants than that determined by the 1:1 Langmuir binding model. Our investigation showed that the equilibrium dissociation constants (KD) determined by a two-step binding model and the 1:1 Langmuir model could differ significantly. The reported findings can facilitate a deeper understanding of antigen-antibody immune complex formation steps and can open a new way for the evaluation of antibody affinity towards corresponding antigens.
Keywords: QCM–D; SARS-CoV-2; affinity interaction; antigen–antibody binding kinetics; immune complex formation; spike protein.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Fenwick C., Croxatto A., Coste A.T., Pojer F., André C., Pellaton C., Farina A., Campos J., Hacker D., Lau K., et al. Changes in SARS-CoV-2 spike versus nucleoprotein antibody responses impact the estimates of infections in population-based seroprevalence studies. J. Virol. 2021;95:e01828-20. doi: 10.1128/JVI.01828-20. - DOI - PMC - PubMed
-
- Poh C.M., Carissimo G., Wang B., Amrun S.N., Lee C.Y., Chee R.S., Fong S., Yeo N.K., Lee W., Torres-ruesta A., et al. Two linear epitopes on the SARS-CoV-2 spike protein that elicit neutralising antibodies in COVID-19 patients. Nat. Commun. 2020;11:2806. doi: 10.1038/s41467-020-16638-2. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

