THBS1-Mediated Degradation of Collagen via the PI3K/AKT Pathway Facilitates the Metastasis and Poor Prognosis of OSCC
- PMID: 37686118
- PMCID: PMC10488045
- DOI: 10.3390/ijms241713312
THBS1-Mediated Degradation of Collagen via the PI3K/AKT Pathway Facilitates the Metastasis and Poor Prognosis of OSCC
Abstract
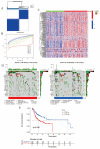
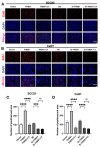
Oral squamous cell carcinoma (OSCC) is a prevalent form of malignant tumor, characterized by a persistently high incidence and mortality rate. The extracellular matrix (ECM) plays a crucial role in the initiation, progression, and diverse biological behaviors of OSCC, facilitated by mechanisms such as providing structural support, promoting cell migration and invasion, regulating cell morphology, and modulating signal transduction. This study investigated the involvement of ECM-related genes, particularly THBS1, in the prognosis and cellular behavior of OSCC. The analysis of ECM-related gene data from OSCC samples identified 165 differentially expressed genes forming two clusters with distinct prognostic outcomes. Seventeen ECM-related genes showed a significant correlation with survival. Experimental methods were employed to demonstrate the impact of THBS1 on proliferation, migration, invasion, and ECM degradation in OSCC cells. A risk-prediction model utilizing four differentially prognostic genes demonstrated significant predictive value in overall survival. THBS1 exhibited enrichment of the PI3K/AKT pathway, indicating its potential role in modulating OSCC. In conclusion, this study observed and verified that ECM-related genes, particularly THBS1, have the potential to influence the prognosis, biological behavior, and immunotherapy of OSCC. These findings hold significant implications for enhancing survival outcomes and providing guidance for precise treatment of OSCC.
Keywords: collagen degradation; extracellular matrix (ECM); immunotherapy; oral squamous cell carcinoma (OSCC); prognosis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
THBS1 is induced by TGFB1 in the cancer stroma and promotes invasion of oral squamous cell carcinoma.J Oral Pathol Med. 2016 Nov;45(10):730-739. doi: 10.1111/jop.12430. Epub 2016 Feb 5. J Oral Pathol Med. 2016. PMID: 26850833
-
Exosomal long noncoding RNAs MAGI2-AS3 and CCDC144NL-AS1 in oral squamous cell carcinoma development via the PI3K-AKT-mTOR signaling pathway.Pathol Res Pract. 2022 Dec;240:154219. doi: 10.1016/j.prp.2022.154219. Epub 2022 Nov 13. Pathol Res Pract. 2022. PMID: 36401978
-
FERMT1 knockdown inhibits oral squamous cell carcinoma cell epithelial-mesenchymal transition by inactivating the PI3K/AKT signaling pathway.BMC Oral Health. 2021 Nov 23;21(1):598. doi: 10.1186/s12903-021-01955-9. BMC Oral Health. 2021. PMID: 34814915 Free PMC article.
-
Abnormal downregulation of 10-formyltetrahydrofolate dehydrogenase promotes the progression of oral squamous cell carcinoma by activating PI3K/Akt/Rb pathway.Cancer Med. 2023 Mar;12(5):5781-5797. doi: 10.1002/cam4.5327. Epub 2022 Nov 6. Cancer Med. 2023. PMID: 36336972 Free PMC article.
-
Matrix-based molecular mechanisms, targeting and diagnostics in oral squamous cell carcinoma.IUBMB Life. 2024 Jul;76(7):368-382. doi: 10.1002/iub.2803. Epub 2024 Jan 2. IUBMB Life. 2024. PMID: 38168122 Review.
Cited by
-
Engineering a dynamic extracellular matrix using thrombospondin-1 to propel hepatocyte organoids reprogramming and improve mouse liver regeneration post-transplantation.Mater Today Bio. 2025 Mar 25;32:101700. doi: 10.1016/j.mtbio.2025.101700. eCollection 2025 Jun. Mater Today Bio. 2025. PMID: 40225139 Free PMC article.
-
LncRNA HOTAIR Interaction With WTAP Promotes m6A Methyltransferase Complex Assembly and Posterior Capsule Opacification Formation by Increasing THBS1.Invest Ophthalmol Vis Sci. 2025 May 1;66(5):20. doi: 10.1167/iovs.66.5.20. Invest Ophthalmol Vis Sci. 2025. PMID: 40341312 Free PMC article.
-
Harnessing tetrahedral framework nucleic acids for enhanced delivery of microRNA-149-3p: A new frontier in oral squamous cell carcinoma therapy.Cell Prolif. 2024 Aug;57(8):e13637. doi: 10.1111/cpr.13637. Epub 2024 Apr 26. Cell Prolif. 2024. PMID: 38671577 Free PMC article.
-
Infection with COVID-19 promotes the progression of pancreatic cancer through the PI3K-AKT signaling pathway.Discov Oncol. 2023 Dec 8;14(1):225. doi: 10.1007/s12672-023-00842-9. Discov Oncol. 2023. PMID: 38063927 Free PMC article.
-
Dysregulated PI3K/AKT signaling in oral squamous cell carcinoma: The tumor microenvironment and epigenetic modifiers as key drivers.Oncol Res. 2025 Jul 18;33(8):1835-1860. doi: 10.32604/or.2025.064010. eCollection 2025. Oncol Res. 2025. PMID: 40746882 Free PMC article. Review.
References
-
- Magnano S., Barroeta P.H., Duffy R., O’Sullivan J., Zisterer D.M. Cisplatin induces autophagy-associated apoptosis in human oral squamous cell carcinoma (OSCC) me-diated in part through reactive oxygen species. Toxicol. Appl. Pharmacol. 2021;427:115646. doi: 10.1016/j.taap.2021.115646. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

