Insight into Tetramolecular DNA G-Quadruplexes Associated with ALS and FTLD: Cation Interactions and Formation of Higher-Ordered Structure
- PMID: 37686239
- PMCID: PMC10487854
- DOI: 10.3390/ijms241713437
Insight into Tetramolecular DNA G-Quadruplexes Associated with ALS and FTLD: Cation Interactions and Formation of Higher-Ordered Structure
Abstract
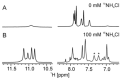
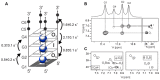
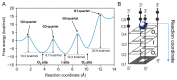
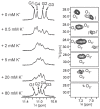
The G4C2 hexanucleotide repeat expansion in the c9orf72 gene is a major genetic cause of familial amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD), with the formation of G-quadruplexes directly linked to the development of these diseases. Cations play a crucial role in the formation and structure of G-quadruplexes. In this study, we investigated the impact of biologically relevant potassium ions on G-quadruplex structures and utilized 15N-labeled ammonium cations as a substitute for K+ ions to gain further insights into cation binding and exchange dynamics. Through nuclear magnetic resonance spectroscopy and molecular dynamics simulations, we demonstrate that the single d(G4C2) repeat, in the presence of 15NH4+ ions, adopts a tetramolecular G-quadruplex with an all-syn quartet at the 5'-end. The movement of 15NH4+ ions through the central channel of the G-quadruplex, as well as to the bulk solution, is governed by the vacant cation binding site, in addition to the all-syn quartet at the 5'-end. Furthermore, the addition of K+ ions to G-quadruplexes folded in the presence of 15NH4+ ions induces stacking of G-quadruplexes via their 5'-end G-quartets, leading to the formation of stable higher-ordered species.
Keywords: ALS; DNA; FTLD; NMR; c9orf7; cations; quadruplex.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Monchaud D. Biological Relevance & Therapeutic Applications of DNA-& RNA-Quadruplexes. Future Science; London, UK: 2015.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

