Dynamics of the Apo µ-Opioid Receptor in Complex with Gi Protein
- PMID: 37686252
- PMCID: PMC10487971
- DOI: 10.3390/ijms241713430
Dynamics of the Apo µ-Opioid Receptor in Complex with Gi Protein
Abstract
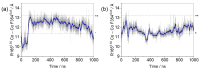
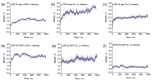
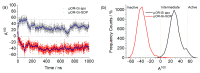
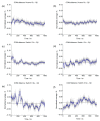
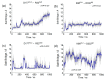
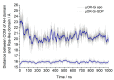
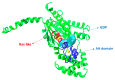
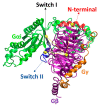
Opioid receptors, particularly the µ-opioid receptor (μOR), play a pivotal role in mediating the analgesic and addictive effects of opioid drugs. G protein signaling is an important pathway of μOR function, usually associated with painkilling effects. However, the molecular mechanisms underlying the interaction between the μOR and G protein remain poorly understood. In this study, we employed classical all-atom molecular dynamics simulations to investigate the structural changes occurring with the μOR-G protein complex under two different conditions: with the G protein in the apo form (open) and with the GDP bound G protein (closed, holo form). The receptor was in the apo form and active conformation in both cases, and the simulation time comprised 1µs for each system. In order to assess the effect of the G protein coupling on the receptor activation state, three parameters were monitored: the correlation of the distance between TM3 and TM6 and the RMSD of the NPxxYA motif; the universal activation index (A100); and the χ2 dihedral distribution of residue W2936.48. When complexed with the open G protein, receptor conformations with intermediate activation state prevailed throughout the molecular dynamics, whereas in the condition with the closed G protein, mostly inactive conformations of the receptor were observed. The major effect of the G protein in the receptor conformation comes from a steric hindrance involving an intracellular loop of the receptor and a β-sheet region of the G protein. This suggests that G-protein precoupling is essential for receptor activation, but this fact is not sufficient for complete receptor activation.
Keywords: G protein signaling; Guanosine Di-phosphate (GDP); molecular dynamics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures










References
-
- Okude J., Ueda T., Kofuku Y., Sato M., Nobuyama N., Kondo K., Shiraishi Y., Mizumura T., Onishi K., Natsume M., et al. Identification of a Conformational Equilibrium That Determines the Efficacy and Functional Selectivity of the μ-Opioid Receptor. Angew. Chem. Int. Ed. 2015;54:15771–15776. doi: 10.1002/anie.201508794. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

