RNA-Seq and Comparative Transcriptomic Analyses of Asian Soybean Rust Resistant and Susceptible Soybean Genotypes Provide Insights into Identifying Disease Resistance Genes
- PMID: 37686258
- PMCID: PMC10487414
- DOI: 10.3390/ijms241713450
RNA-Seq and Comparative Transcriptomic Analyses of Asian Soybean Rust Resistant and Susceptible Soybean Genotypes Provide Insights into Identifying Disease Resistance Genes
Abstract
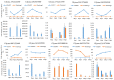
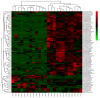
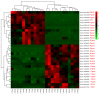
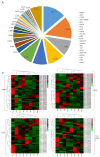
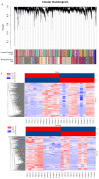
Asian soybean rust (ASR), caused by Phakopsora pachyrhizi, is one of the most destructive foliar diseases that affect soybeans. Developing resistant cultivars is the most cost-effective, environmentally friendly, and easy strategy for controlling the disease. However, the current understanding of the mechanisms underlying soybean resistance to P. pachyrhizi remains limited, which poses a significant challenge in devising effective control strategies. In this study, comparative transcriptomic profiling using one resistant genotype and one susceptible genotype was performed under infected and control conditions to understand the regulatory network operating between soybean and P. pachyrhizi. RNA-Seq analysis identified a total of 6540 differentially expressed genes (DEGs), which were shared by all four genotypes. The DEGs are involved in defense responses, stress responses, stimulus responses, flavonoid metabolism, and biosynthesis after infection with P. pachyrhizi. A total of 25,377 genes were divided into 33 modules using weighted gene co-expression network analysis (WGCNA). Two modules were significantly associated with pathogen defense. The DEGs were mainly enriched in RNA processing, plant-type hypersensitive response, negative regulation of cell growth, and a programmed cell death process. In conclusion, these results will provide an important resource for mining resistant genes to P. pachyrhizi infection and valuable resources to potentially pyramid quantitative resistance loci for improving soybean germplasm.
Keywords: Asian soybean rust; RNA-seq; WGCNA; soybean.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Akamatsu H., Yamanaka N., Soares R.M., Ivancovich A., Kato M. Pathogenic variation of south American Phakopsora pachyrhizi populations isolated from soybeans from 2010 to 2015. Jpn. Agric. Res. Q. Jarq. 2017;51:221–232. doi: 10.6090/jarq.51.221. - DOI
-
- Godoy C., de Freitas Bueno A., Gazziero D. Brazilian soybean pest management and threats to its sustainability. Outlooks Pest Manag. 2015;26:113–117. doi: 10.1564/v26_jun_06. - DOI
-
- Vuong T.D., Walker D.R., Nguyen B.T., Nguyen T.T., Dinh H.X., Hyten D.L., Cregan P.B., Sleper D.A., Lee J.D., Shannon J.G., et al. Molecular characterization of resistance to soybean rust (Phakopsora pachyrhizi Syd. & Syd.) in soybean cultivar DT 2000 (PI 635999) PLoS ONE. 2016;11:e0164493. doi: 10.1371/journal.pone.0164493. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

