Prenatal Alcohol Exposure Impairs the Placenta-Cortex Transcriptomic Signature, Leading to Dysregulation of Angiogenic Pathways
- PMID: 37686296
- PMCID: PMC10488081
- DOI: 10.3390/ijms241713484
Prenatal Alcohol Exposure Impairs the Placenta-Cortex Transcriptomic Signature, Leading to Dysregulation of Angiogenic Pathways
Abstract
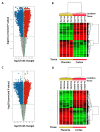
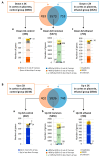
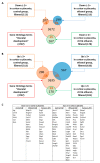
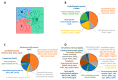
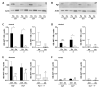
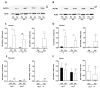
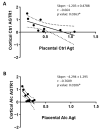
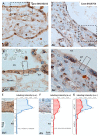
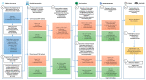
Although alcohol consumption during pregnancy is a major cause of behavioral and learning disabilities, most FASD infants are late- or even misdiagnosed due to clinician's difficulties achieving early detection of alcohol-induced neurodevelopmental impairments. Neuroplacentology has emerged as a new field of research focusing on the role of the placenta in fetal brain development. Several studies have reported that prenatal alcohol exposure (PAE) dysregulates a functional placenta-cortex axis, which is involved in the control of angiogenesis and leads to neurovascular-related defects. However, these studies were focused on PlGF, a pro-angiogenic factor. The aim of the present study is to provide the first transcriptomic "placenta-cortex" signature of the effects of PAE on fetal angiogenesis. Whole mouse genome microarrays of paired placentas and cortices were performed to establish the transcriptomic inter-organ "placenta-cortex" signature in control and PAE groups at gestational day 20. Genespring comparison of the control and PAE signatures revealed that 895 and 1501 genes were only detected in one of two placenta-cortex expression profiles, respectively. Gene ontology analysis indicated that 107 of these genes were associated with vascular development, and String protein-protein interaction analysis showed that they were associated with three functional clusters. PANTHER functional classification analysis indicated that "intercellular communication" was a significantly enriched biological process, and 27 genes were encoded for neuroactive ligand/receptors interactors. Protein validation experiments involving Western blot for one ligand-receptor couple (Agt/AGTR1/2) confirmed the transcriptomic data, and Pearson statistical analysis of paired placentas and fetal cortices revealed a negative correlation between placental Atg and cortical AGTR1, which was significantly impacted by PAE. In humans, a comparison of a 38WG control placenta with a 36WG alcohol-exposed placenta revealed low Agt immunolabeling in the syncytiotrophoblast layer of the alcohol case. In conclusion, this study establishes the first transcriptomic placenta-cortex signature of a developing mouse. The data show that PAE markedly unbalances this inter-organ signature; in particular, several ligands and/or receptors involved in the control of angiogenesis. These data support that PAE modifies the existing communication between the two organs and opens new research avenues regarding the impact of placental dysfunction on the neurovascular development of fetuses. Such a signature would present a clinical value for early diagnosis of brain defects in FASD.
Keywords: FASD; diagnosis; neurodevelopment; neuroplacentology; neurovascular.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
PLGF, a placental marker of fetal brain defects after in utero alcohol exposure.Acta Neuropathol Commun. 2017 Jun 6;5(1):44. doi: 10.1186/s40478-017-0444-6. Acta Neuropathol Commun. 2017. PMID: 28587682 Free PMC article.
-
Chromatin modifier developmental pluripotency associated factor 4 (DPPA4) is a candidate gene for alcohol-induced developmental disorders.BMC Med. 2022 Dec 30;20(1):495. doi: 10.1186/s12916-022-02699-1. BMC Med. 2022. PMID: 36581877 Free PMC article.
-
RNA-seq analysis reveals prenatal alcohol exposure is associated with placental inflammatory cells and gene expression.Gene. 2024 Feb 5;894:147951. doi: 10.1016/j.gene.2023.147951. Epub 2023 Oct 31. Gene. 2024. PMID: 37918548 Free PMC article.
-
Long-term alterations to DNA methylation as a biomarker of prenatal alcohol exposure: From mouse models to human children with fetal alcohol spectrum disorders.Alcohol. 2017 May;60:67-75. doi: 10.1016/j.alcohol.2016.11.009. Epub 2016 Nov 22. Alcohol. 2017. PMID: 28187949 Review.
-
Potential roles of imprinted genes in the teratogenic effects of alcohol on the placenta, somatic growth, and the developing brain.Exp Neurol. 2022 Jan;347:113919. doi: 10.1016/j.expneurol.2021.113919. Epub 2021 Nov 6. Exp Neurol. 2022. PMID: 34752786 Review.
Cited by
-
Prenatal alcohol exposure is associated with altered feto-placental blood flow and sex-specific placental changes.JCI Insight. 2025 Feb 10;10(3):e186096. doi: 10.1172/jci.insight.186096. JCI Insight. 2025. PMID: 39927463 Free PMC article.
-
Expression of placental CD146 is dysregulated by prenatal alcohol exposure and contributes in cortical vasculature development and positioning of vessel-associated oligodendrocytes.Front Cell Neurosci. 2024 Jan 10;17:1294746. doi: 10.3389/fncel.2023.1294746. eCollection 2023. Front Cell Neurosci. 2024. PMID: 38269113 Free PMC article.
-
Nonconceptus Mechanisms of Prenatal Alcohol Exposure That Disrupt Embryo-Fetal Development: An Integrative View.Alcohol Res. 2025 Jul 16;45(1):07. doi: 10.35946/arcr.v45.1.07. eCollection 2025. Alcohol Res. 2025. PMID: 40692661 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

