Oligomeric State of β-Coronavirus Non-Structural Protein 10 Stimulators Studied by Small Angle X-ray Scattering
- PMID: 37686452
- PMCID: PMC10563069
- DOI: 10.3390/ijms241713649
Oligomeric State of β-Coronavirus Non-Structural Protein 10 Stimulators Studied by Small Angle X-ray Scattering
Abstract
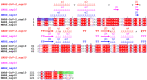
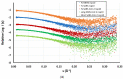
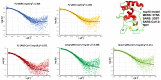
The β-coronavirus family, encompassing Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), Severe Acute Respiratory Syndrome Coronavirus (SARS), and Middle East Respiratory Syndrome Coronavirus (MERS), has triggered pandemics within the last two decades. With the possibility of future pandemics, studying the coronavirus family members is necessary to improve knowledge and treatment. These viruses possess 16 non-structural proteins, many of which play crucial roles in viral replication and in other vital functions. One such vital protein is non-structural protein 10 (nsp10), acting as a pivotal stimulator of nsp14 and nsp16, thereby influencing RNA proofreading and viral RNA cap formation. Studying nsp10 of pathogenic coronaviruses is central to unraveling its multifunctional roles. Our study involves the biochemical and biophysical characterisation of full-length nsp10 from MERS, SARS and SARS-CoV-2. To elucidate their oligomeric state, we employed a combination of Multi-detection Size exclusion chromatography (Multi-detection SEC) with multi-angle static light scattering (MALS) and small angle X-ray scattering (SAXS) techniques. Our findings reveal that full-length nsp10s primarily exist as monomers in solution, while truncated versions tend to oligomerise. SAXS experiments reveal a globular shape for nsp10, a trait conserved in all three coronaviruses, although MERS nsp10, diverges most from SARS and SARS-CoV-2 nsp10s. In summary, unbound nsp10 proteins from SARS, MERS, and SARS-CoV-2 exhibit a globular and predominantly monomeric state in solution.
Keywords: COVID-19; SARS-CoV-2; SAXS; conformational changes; non-structural proteins; nsp10; oligomeric state.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
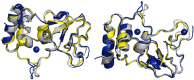



Similar articles
-
Structural and functional insights into the 2'-O-methyltransferase of SARS-CoV-2.Virol Sin. 2024 Aug;39(4):619-631. doi: 10.1016/j.virs.2024.07.001. Epub 2024 Jul 3. Virol Sin. 2024. PMID: 38969340 Free PMC article.
-
Binding of the Methyl Donor S-Adenosyl-l-Methionine to Middle East Respiratory Syndrome Coronavirus 2'-O-Methyltransferase nsp16 Promotes Recruitment of the Allosteric Activator nsp10.J Virol. 2017 Feb 14;91(5):e02217-16. doi: 10.1128/JVI.02217-16. Print 2017 Mar 1. J Virol. 2017. PMID: 28031370 Free PMC article.
-
SS148 and WZ16 inhibit the activities of nsp10-nsp16 complexes from all seven human pathogenic coronaviruses.Biochim Biophys Acta Gen Subj. 2023 Apr;1867(4):130319. doi: 10.1016/j.bbagen.2023.130319. Epub 2023 Feb 9. Biochim Biophys Acta Gen Subj. 2023. PMID: 36764586 Free PMC article.
-
Human and novel coronavirus infections in children: a review.Paediatr Int Child Health. 2021 Feb;41(1):36-55. doi: 10.1080/20469047.2020.1781356. Epub 2020 Jun 25. Paediatr Int Child Health. 2021. PMID: 32584199 Review.
-
From SARS and MERS to COVID-19: a brief summary and comparison of severe acute respiratory infections caused by three highly pathogenic human coronaviruses.Respir Res. 2020 Aug 27;21(1):224. doi: 10.1186/s12931-020-01479-w. Respir Res. 2020. PMID: 32854739 Free PMC article.
References
-
- World Health Organization Consensus Document on the Epidemiology of Severe Acute Respiratory Syndrome (SARS) 2003. [(accessed on 1 May 2023)]. Available online: https://www.who.int/publications/i/item/consensus-document-on-the-epidem...
-
- World Health Organization Middle East Respiratory Syndrome Coronavirus (MERS-CoV)—Saudi Arabia. 2022. [(accessed on 8 May 2023)]. Available online: https://www.who.int/emergencies/disease-outbreak-news/item/2022-DON363.
-
- World Health Organization Coronavirus Disease (COVID-19) Pandemic. 2023. [(accessed on 8 May 2023)]. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

