11p15 Epimutations in Pediatric Embryonic Tumors: Insights from a Methylome Analysis
- PMID: 37686532
- PMCID: PMC10486592
- DOI: 10.3390/cancers15174256
11p15 Epimutations in Pediatric Embryonic Tumors: Insights from a Methylome Analysis
Abstract
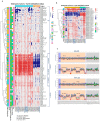
Embryonic tumors share few recurrent mutations, suggesting that other mechanisms, such as aberrant DNA methylation, play a prominent role in their development. The loss of imprinting (LOI) at the chromosome region 11p15 is the germline alteration behind Beckwith-Wiedemann syndrome that results in an increased risk of developing several embryonic tumors. This study analyzed the methylome, using EPIC Beadchip arrays from 99 sporadic embryonic tumors. Among these tumors, 46.5% and 14.6% presented alterations at imprinted control regions (ICRs) 1 and 2, respectively. Based on the methylation levels of ICR1 and ICR2, four clusters formed with distinct methylation patterns, mostly for medulloblastomas (ICR1 loss of methylation (LOM)), Wilms tumors, and hepatoblastomas (ICR1 gain of methylation (GOM), with or without ICR2 LOM). To validate the results, the methylation status of 29 cases was assessed with MS-MLPA, and a high level of agreement was found between both methodologies: 93% for ICR1 and 79% for ICR2. The MS-MLPA results indicate that 15 (51.7%) had ICR1 GOM and 11 (37.9%) had ICR2 LOM. To further validate our findings, the ICR1 methylation status was characterized via digital PCR (dPCR) in cell-free DNA (cfDNA) extracted from peripheral blood. At diagnosis, we detected alterations in the methylation levels of ICR1 in 62% of the cases, with an agreement of 76% between the tumor tissue (MS-MLPA) and cfDNA methods. Among the disagreements, the dPCR was able to detect ICR1 methylation level changes presented at heterogeneous levels in the tumor tissue, which were detected only in the methylome analysis. This study highlights the prevalence of 11p15 methylation status in sporadic embryonic tumors, with differences relating to methylation levels (gain or loss), location (ICR1 or ICR2), and tumor types (medulloblastomas, Wilms tumors, and hepatoblastomas).
Keywords: 11p15; DNA methylation; ICR1; ICR2; Wilms tumor; embryonic tumors; medulloblastoma.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources

