Human Aldehyde Dehydrogenases: A Superfamily of Similar Yet Different Proteins Highly Related to Cancer
- PMID: 37686694
- PMCID: PMC10650815
- DOI: 10.3390/cancers15174419
Human Aldehyde Dehydrogenases: A Superfamily of Similar Yet Different Proteins Highly Related to Cancer
Abstract
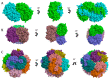
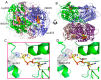
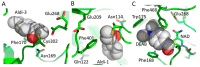
The superfamily of human aldehyde dehydrogenases (hALDHs) consists of 19 isoenzymes which are critical for several physiological and biosynthetic processes and play a major role in the organism's detoxification via the NAD(P) dependent oxidation of numerous endogenous and exogenous aldehyde substrates to their corresponding carboxylic acids. Over the last decades, ALDHs have been the subject of several studies as it was revealed that their differential expression patterns in various cancer types are associated either with carcinogenesis or promotion of cell survival. Here, we attempt to provide a thorough review of hALDHs' diverse functions and 3D structures with particular emphasis on their role in cancer pathology and resistance to chemotherapy. We are especially interested in findings regarding the association of structural features and their changes with effects on enzymes' functionalities. Moreover, we provide an updated outline of the hALDHs inhibitors utilized in experimental or clinical settings for cancer therapy. Overall, this review aims to provide a better understanding of the impact of ALDHs in cancer pathology and therapy from a structural perspective.
Keywords: 3D structure; aldehyde dehydrogenases (ALDHs); cancer; chemotherapy resistance; crystallins; inhibitors; post-translational modifications; quaternary association; topology.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures
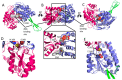
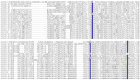
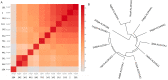
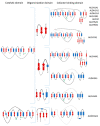




References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

