Sanguisorba officinalis L. Ameliorates Hepatic Steatosis and Fibrosis by Modulating Oxidative Stress, Fatty Acid Oxidation, and Gut Microbiota in CDAHFD-Induced Mice
- PMID: 37686810
- PMCID: PMC10490207
- DOI: 10.3390/nu15173779
Sanguisorba officinalis L. Ameliorates Hepatic Steatosis and Fibrosis by Modulating Oxidative Stress, Fatty Acid Oxidation, and Gut Microbiota in CDAHFD-Induced Mice
Abstract
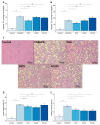
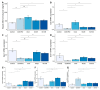
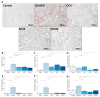
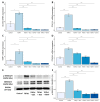
Non-alcoholic fatty liver disease (NAFLD) is a leading cause of chronic liver diseases and encompasses non-alcoholic steatosis, steatohepatitis, and fibrosis. Sanguisorba officinalis L. (SO) roots have traditionally been used for their antioxidant properties and have beneficial effects on metabolic disorders, including diabetes and obesity. However, its effects on hepatic steatosis and fibrosis remain unclear. In this study, we explored the effects of a 95% ethanolic SO extract (SOEE) on NAFLD and fibrosis in vivo and in vitro. The SOEE was orally administered to C57BL/6J mice fed a choline-deficient, L-amino-acid-defined, high-fat diet for 10 weeks. The SOEE inhibited hepatic steatosis by modulating hepatic malondialdehyde levels and the expression of oxidative stress-associated genes, regulating fatty-acid-oxidation-related genes, and inhibiting the expression of genes that are responsible for fibrosis. The SOEE suppressed the deposition of extracellular matrix hydroxyproline and mRNA expression of fibrosis-associated genes. The SOEE decreased the expression of fibrosis-related genes in vitro by inhibiting SMAD2/3 phosphorylation. Furthermore, the SOEE restored the gut microbial diversity and modulated specific bacterial genera associated with NAFLD and fibrosis. This study suggests that SOEE might be the potential candidate for inhibiting hepatic steatosis and fibrosis by modulating oxidative stress, fatty acid oxidation, and gut microbiota composition.
Keywords: L-amino acid-defined; Sanguisorba officinalis L.; choline-deficient; fibrosis; gut microbiota; high-fat diet (CDAHFD); steatosis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Riazi K., Azhari H., Charette J.H., Underwood F.E., King J.A., Afshar E.E., Swain M.G., Congly S.E., Kaplan G.G., Shaheen A.-A. The prevalence and incidence of NAFLD worldwide: A systematic review and meta-analysis. Lancet Gastroenterol. Hepatol. 2022;7:851–861. doi: 10.1016/S2468-1253(22)00165-0. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

