Epigenomic signature of accelerated ageing in progeroid Cockayne syndrome
- PMID: 37688320
- PMCID: PMC10577576
- DOI: 10.1111/acel.13959
Epigenomic signature of accelerated ageing in progeroid Cockayne syndrome
Abstract
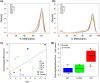
Cockayne syndrome (CS) and UV-sensitive syndrome (UVSS) are rare genetic disorders caused by mutation of the DNA repair and multifunctional CSA or CSB protein, but only CS patients display a progeroid and neurodegenerative phenotype, providing a unique conceptual and experimental paradigm. As DNA methylation (DNAm) remodelling is a major ageing marker, we performed genome-wide analysis of DNAm of fibroblasts from healthy, UVSS and CS individuals. Differential analysis highlighted a CS-specific epigenomic signature (progeroid-related; not present in UVSS) enriched in three categories: developmental transcription factors, ion/neurotransmitter membrane transporters and synaptic neuro-developmental genes. A large fraction of CS-specific DNAm changes were associated with expression changes in CS samples, including in previously reported post-mortem cerebella. The progeroid phenotype of CS was further supported by epigenomic hallmarks of ageing: the prediction of DNAm of repetitive elements suggested an hypomethylation of Alu sequences in CS, and the epigenetic clock returned a marked increase in CS biological age respect to healthy and UVSS cells. The epigenomic remodelling of accelerated ageing in CS displayed both commonalities and differences with other progeroid diseases and regular ageing. CS shared DNAm changes with normal ageing more than other progeroid diseases do, and included genes functionally validated for regular ageing. Collectively, our results support the existence of an epigenomic basis of accelerated ageing in CS and unveil new genes and pathways that are potentially associated with the progeroid/degenerative phenotype.
Keywords: Cockayne syndrome; DNA methylation; UV-sensitive syndrome; ageing; epigenetic clock; progeroid diseases.
© 2023 The Authors. Aging Cell published by Anatomical Society and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they have no conflict of interests.
Figures



References
-
- Bacalini, M. G. , Boattini, A. , Gentilini, D. , Giampieri, E. , Pirazzini, C. , Giuliani, C. , Fontanesi, E. , Remondini, D. , Capri, M. , del Rio, A. , Luiselli, D. , Vitale, G. , Mari, D. , Castellani, G. , di Blasio, A. M. , Salvioli, S. , Franceschi, C. , & Garagnani, P. (2015). A meta‐analysis on age‐associated changes in blood DNA methylation: Results from an original analysis pipeline for infinium 450k data. Aging (Albany NY), 7(2), 97–109. - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

