Germline pathogenic variants in neuroblastoma patients are enriched in BARD1 and predict worse survival
- PMID: 37688579
- PMCID: PMC10777667
- DOI: 10.1093/jnci/djad183
Germline pathogenic variants in neuroblastoma patients are enriched in BARD1 and predict worse survival
Abstract
Background: Neuroblastoma is an embryonal cancer of the developing sympathetic nervous system. The genetic contribution of rare pathogenic or likely pathogenic germline variants in patients without a family history remains unclear.
Methods: Germline DNA sequencing was performed on 786 neuroblastoma patients. The frequency of rare cancer predisposition gene pathogenic or likely pathogenic variants in patients was compared with 2 cancer-free control cohorts. Matched tumor DNA sequencing was evaluated for second hits, and germline DNA array data from 5585 neuroblastoma patients and 23 505 cancer-free control children were analyzed to identify rare germline copy number variants. Patients with germline pathogenic or likely pathogenic variants were compared with those without to test for association with clinical characteristics, tumor features, and survival.
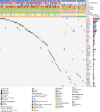
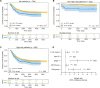
Results: We observed 116 pathogenic or likely pathogenic variants involving 13.9% (109 of 786) of neuroblastoma patients, representing a statistically significant excess burden compared with cancer-free participants (odds ratio [OR] = 1.60, 95% confidence interval [CI] = 1.27 to 2.00). BARD1 harbored the most statistically significant enrichment of pathogenic or likely pathogenic variants (OR = 32.30, 95% CI = 6.44 to 310.35). Rare germline copy number variants disrupting BARD1 were identified in patients but absent in cancer-free participants (OR = 29.47, 95% CI = 1.52 to 570.70). Patients harboring a germline pathogenic or likely pathogenic variant had a worse overall survival compared with those without (P = 8.6 x 10-3).
Conclusions: BARD1 is an important neuroblastoma predisposition gene harboring both common and rare germline pathogenic or likely pathogenic variations. The presence of any germline pathogenic or likely pathogenic variant in a cancer predisposition gene was independently predictive of worse overall survival. As centers move toward paired tumor-normal sequencing at diagnosis, efforts should be made to centralize data and provide an infrastructure to support cooperative longitudinal prospective studies of germline pathogenic variation.
© The Author(s) 2023. Published by Oxford University Press. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Conflict of interest statement
The authors have no disclosures.
Figures


Update of
-
Germline pathogenic variants in 786 neuroblastoma patients.medRxiv [Preprint]. 2023 Jan 25:2023.01.23.23284864. doi: 10.1101/2023.01.23.23284864. medRxiv. 2023. Update in: J Natl Cancer Inst. 2024 Jan 10;116(1):149-159. doi: 10.1093/jnci/djad183. PMID: 36747619 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
- R03 CA230366/CA/NCI NIH HHS/United States
- R01 CA237562/CA/NCI NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- K08 CA215312/CA/NCI NIH HHS/United States
- T32 CA151022/CA/NCI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R35 CA220500/CA/NCI NIH HHS/United States
- R01 CA204974/CA/NCI NIH HHS/United States
- HHSN261200800001E/NH/NIH HHS/United States
- K08 CA230223/CA/NCI NIH HHS/United States
- U10 CA098543/CA/NCI NIH HHS/United States
- HHSN261200800001C/RC/CCR NIH HHS/United States
- HHSN261200800001E/CA/NCI NIH HHS/United States
- R01CA204974/NH/NIH HHS/United States
- PST-07-16/DRCRF/Damon Runyon Cancer Research Foundation/United States

