Novel findings about the mode of action of the antifungal protein PeAfpA against Saccharomyces cerevisiae
- PMID: 37688596
- PMCID: PMC10589166
- DOI: 10.1007/s00253-023-12749-0
Novel findings about the mode of action of the antifungal protein PeAfpA against Saccharomyces cerevisiae
Abstract
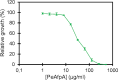
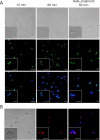
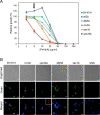
Antifungal proteins (AFPs) from filamentous fungi offer the potential to control fungal infections that threaten human health and food safety. AFPs exhibit broad antifungal spectra against harmful fungi, but limited knowledge of their killing mechanism hinders their potential applicability. PeAfpA from Penicillium expansum shows strong antifungal potency against plant and human fungal pathogens and stands above other AFPs for being active against the yeast Saccharomyces cerevisiae. We took advantage of this and used a model laboratory strain of S. cerevisiae to gain insight into the mode of action of PeAfpA by combining (i) transcriptional profiling, (ii) PeAfpA sensitivity analyses of deletion mutants available in the S. cerevisiae genomic deletion collection and (iii) cell biology studies using confocal microscopy. Results highlighted and confirmed the role of the yeast cell wall (CW) in the interaction with PeAfpA, which can be internalized through both energy-dependent and independent mechanisms. The combined results also suggest an active role of the CW integrity (CWI) pathway and the cAMP-PKA signalling in the PeAfpA killing mechanism. Besides, our studies revealed the involvement of phosphatidylinositol metabolism and the participation of ROX3, which codes for the subunit 19 of the RNA polymerase II mediator complex, in the yeast defence strategy. In conclusion, our study provides clues about both the killing mechanism of PeAfpA and the fungus defence strategies against the protein, suggesting also targets for the development of new antifungals. KEY POINTS: • PeAfpA is a cell-penetrating protein with inhibitory activity against S. cerevisiae. • The CW integrity (CWI) pathway is a key player in the PeAfpA killing mechanism. • Phosphatidylinositol metabolism and ROX3 are involved in the yeast defence strategy.
Keywords: Antifungal proteins (AFPs); Cell wall integrity; Cell-penetrating protein; Phosphatidylinositol metabolism; Transcriptomics; Yeast deletion collection.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Atanasova L, Moreno-Ruiz D, Grünwald-Gruber C, Hell V, Zeilinger S (2021) The GPI-anchored GH76 protein Dfg5 affects hyphal morphology and osmoregulation in the mycoparasite Trichoderma atroviride and is interconnected with MAPK signaling. Front Microbiol 12:601113. 10.3389/fmicb.2021.601113 - PMC - PubMed
-
- Batta G, Barna T, Gáspári Z, Sándor S, Kövér KE, Binder U, Sarg B, Kaiserer L, Chhillar AK, Eigentler A, Leiter É, Hegedüs N, Pócsi I, Lindner H, Marx F. Functional aspects of the solution structure and dynamics of PAF – a highly-stable antifungal protein from Penicillium chrysogenum. FEBS J. 2009;276(10):2875–2890. doi: 10.1111/j.1742-4658.2009.07011.x. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

