TIMP1 shapes an immunosuppressive microenvironment by regulating anoikis to promote the progression of clear cell renal cell carcinoma
- PMID: 37688768
- PMCID: PMC10522382
- DOI: 10.18632/aging.205005
TIMP1 shapes an immunosuppressive microenvironment by regulating anoikis to promote the progression of clear cell renal cell carcinoma
Abstract
Background: The association between ccRCC and Anoikis remains to be thoroughly investigated.
Methods: Anoikis-related clusters were identified using NMF. To identify prognostic anoikis-related genes (ARGs) and establish an optimal prognostic model, univariate Cox and LASSO regression were employed. The E-MTAB-1980 cohort was utilized for external validation. Multiple algorithms were used to evaluate the immune properties of the model. GO, KEGG and GSVA analyses were employed to analyze biological pathway functions. qRT-PCR was employed to measure RNA levels of specific genes. Cell Counting Kit-8, wound healing, and Transwell chamber assays were performed to determine changes in the proliferative and metastatic abilities of A498 and 786-O cells.
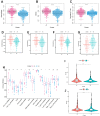
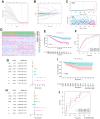
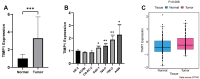
Results: Based on the expression of 21 prognostic ARGs, we constructed anoikis-related clusters with different prognostic and immune characteristics. The cluster A1 showed a worse prognosis, higher infiltration of immunosuppressive cells and enrichment of several oncogenic pathways. We also calculated the Anoikis Index (AI). Patients in high AI group had a worse prognosis, higher infiltration of immunosuppressive cells and higher expression of immunosuppressive checkpoints. TIMP1 exerted a tumor-promoting role in ccRCC and was significantly associated with immunosuppressive cells and checkpoints. The downregulation of TIMP1 negatively regulated ccRCC cell proliferation and metastasis.
Conclusions: ARGs played crucial roles in tumorigenesis and progression and were positively associated with a poor prognosis. AI had great accuracy in predicting the prognosis and immune characteristics of ccRCC patients. TIMP1 was significantly associated with clinicopathological variables and the immunosuppressive microenvironment, which could be exploited to design novel immunotherapies for ccRCC patients.
Keywords: TIMP1; anoikis; anoikis index; ccRCC; tumor microenvironment.
Conflict of interest statement
Figures






References
-
- Li Y, Lih TM, Dhanasekaran SM, Mannan R, Chen L, Cieslik M, Wu Y, Lu RJ, Clark DJ, Kołodziejczak I, Hong R, Chen S, Zhao Y, et al. , and Clinical Proteomic Tumor Analysis Consortium. Histopathologic and proteogenomic heterogeneity reveals features of clear cell renal cell carcinoma aggressiveness. Cancer Cell. 2023; 41:139–63.e17. 10.1016/j.ccell.2022.12.001 - DOI - PMC - PubMed
-
- Yu Z, Lv Y, Su C, Lu W, Zhang R, Li J, Guo B, Yan H, Liu D, Yang Z, Mi H, Mo L, Guo Y, et al. Integrative Single-Cell Analysis Reveals Transcriptional and Epigenetic Regulatory Features of Clear Cell Renal Cell Carcinoma. Cancer Res. 2023; 83:700–19. 10.1158/0008-5472.CAN-22-2224 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

