Screening in serum-derived medium reveals differential response to compounds targeting metabolism
- PMID: 37689063
- PMCID: PMC10581593
- DOI: 10.1016/j.chembiol.2023.08.007
Screening in serum-derived medium reveals differential response to compounds targeting metabolism
Abstract
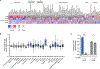
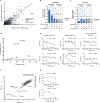
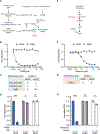
A challenge for screening new anticancer drugs is that efficacy in cell culture models is not always predictive of efficacy in patients. One limitation of standard cell culture is a reliance on non-physiological nutrient levels, which can influence cell metabolism and drug sensitivity. A general assessment of how physiological nutrients affect cancer cell response to small molecule therapies is lacking. To address this, we developed a serum-derived culture medium that supports the proliferation of diverse cancer cell lines and is amenable to high-throughput screening. We screened several small molecule libraries and found that compounds targeting metabolic enzymes were differentially effective in standard compared to serum-derived medium. We exploited the differences in nutrient levels between each medium to understand why medium conditions affected the response of cells to some compounds, illustrating how this approach can be used to screen potential therapeutics and understand how their efficacy is modified by available nutrients.
Keywords: Cancer cell metabolism; Culture media; Drug sensitivity; High-throughput screening; Nutrient environment; Phenotypic drug screening; Physiologic media.
Copyright © 2023 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of interests M.G.V.H. discloses that he is a scientific advisor for Agios Pharmaceuticals, iTeos Therapeutics, Sage Therapeutics, Pretzel Therapeutics, Lime Therapeutics, Faeth Therapeutics, Droia Ventures, and Auron Therapeutics. D.C., S.R., A.F., L.M.G., and D.S.A. are (or were at the time of their involvement with this study) employees of Novartis. P.P.H. has consulted for Auron Therapeutics. I.M.T.G. is a current employee of AstraZeneca. All remaining authors declare no competing interests.
Figures


Update of
-
Screening in serum-derived medium reveals differential response to compounds targeting metabolism.bioRxiv [Preprint]. 2023 Feb 27:2023.02.25.529972. doi: 10.1101/2023.02.25.529972. bioRxiv. 2023. Update in: Cell Chem Biol. 2023 Sep 21;30(9):1156-1168.e7. doi: 10.1016/j.chembiol.2023.08.007. PMID: 36909640 Free PMC article. Updated. Preprint.
References
-
- Davidson SM, Papagiannakopoulos T, Olenchock BA, Heyman JE, Keibler MA, Luengo A, Bauer MR, Jha AK, O’Brien JP, Pierce KA, et al. (2016). Environment Impacts the Metabolic Dependencies of Ras-Driven Non-Small Cell Lung Cancer. Cell Metab. 23, 517–528. 10.1016/j.cmet.2016.01.007. - DOI - PMC - PubMed
-
- Gui DY, Sullivan LB, Luengo A, Hosios AM, Bush LN, Gitego N, Davidson SM, Freinkman E, Thomas CJ, and Vander Heiden MG (2016). Environment Dictates Dependence on Mitochondrial Complex I for NAD+ and Aspartate Production and Determines Cancer Cell Sensitivity to Metformin. Cell Metab. 24, 716–727. 10.1016/j.cmet.2016.09.006. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- KL2 TR002542/TR/NCATS NIH HHS/United States
- U01 CA224348/CA/NCI NIH HHS/United States
- R01 CA259253/CA/NCI NIH HHS/United States
- 110302/Z/15/Z/WT_/Wellcome Trust/United Kingdom
- P30 CA014051/CA/NCI NIH HHS/United States
- T32 GM007753/GM/NIGMS NIH HHS/United States
- R35 CA242379/CA/NCI NIH HHS/United States
- T32 GM144273/GM/NIGMS NIH HHS/United States
- 204845/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- R01 CA208205/CA/NCI NIH HHS/United States
- T32 CA071345/CA/NCI NIH HHS/United States
- MR/T032413/1/MRC_/Medical Research Council/United Kingdom
- P50 CA090381/CA/NCI NIH HHS/United States
- F30 HL156404/HL/NHLBI NIH HHS/United States
- P30 DK040561/DK/NIDDK NIH HHS/United States
- T32 GM007287/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- DH_/Department of Health/United Kingdom
- R01 CA269672/CA/NCI NIH HHS/United States
- U01 CA261842/CA/NCI NIH HHS/United States
- F31 CA271787/CA/NCI NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources

