Expanding the clinical and immunological phenotypes of PAX1-deficient SCID and CID patients
- PMID: 37689091
- PMCID: PMC10958138
- DOI: 10.1016/j.clim.2023.109757
Expanding the clinical and immunological phenotypes of PAX1-deficient SCID and CID patients
Erratum in
-
Corrigendum to "Expanding the clinical and immunological phenotypes of PAX1-deficient SCID and CID patients" [Clinical Immunology 255 (2023) 109757].Clin Immunol. 2023 Nov;256:109799. doi: 10.1016/j.clim.2023.109799. Epub 2023 Oct 14. Clin Immunol. 2023. PMID: 37845128 Free PMC article. No abstract available.
Abstract
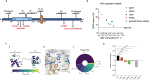
Paired box 1 (PAX1) deficiency has been reported in a small number of patients diagnosed with otofaciocervical syndrome type 2 (OFCS2). We described six new patients who demonstrated variable clinical penetrance. Reduced transcriptional activity of pathogenic variants confirmed partial or complete PAX1 deficiency. Thymic aplasia and hypoplasia were associated with impaired T cell immunity. Corrective treatment was required in 4/6 patients. Hematopoietic stem cell transplantation resulted in poor immune reconstitution with absent naïve T cells, contrasting with the superior recovery of T cell immunity after thymus transplantation. Normal ex vivo differentiation of PAX1-deficient CD34+ cells into mature T cells demonstrated the absence of a hematopoietic cell-intrinsic defect. New overlapping features with DiGeorge syndrome included primary hypoparathyroidism (n = 5) and congenital heart defects (n = 2), in line with PAX1 expression during early embryogenesis. Our results highlight new features of PAX1 deficiency, which are relevant to improving early diagnosis and identifying patients requiring corrective treatment.
Keywords: Hypoparathyroidism; Inborn errors of immunity; Otofaciocervical syndrome; PAX1; SCID; thymus.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest All authors declare no conflict of interest to disclose.
Figures




References
-
- Estefania E., Ramírez-Camacho R., Gomar M., Trinidad A., Arellano B., Garcia-Berrocal J., Verdaguer J., Vilches C. Point mutation of an EYA1-gene splice site in a patient with oto-facio-cervical syndrome. Ann. Hum. Genet. 2006;70:140–144. - PubMed
-
- Pohl E., Aykut A., Beleggia F., Karaca E., Durmaz B., Keupp K., Arslan E., Onay M.P., Yigit G., Özkinay F. A hypofunctional PAX1 mutation causes autosomal recessively inherited otofaciocervical syndrome. Hum. Genet. 2013;132:1311–1320. - PubMed
-
- Paganini I., Sestini R., Capone G., Putignano A., Contini E., Giotti I., Gensini F., Marozza A., Barilaro A., Porfirio B. A novel PAX1 null homozygous mutation in autosomal recessive otofaciocervical syndrome associated with severe combined immunodeficiency. Clin. Genet. 2017;92:664–668. - PubMed
-
- Patil S.J., Das Bhowmik A., Bhat V., Satidevi Vineeth V., Vasudevamurthy R., Dalal A. Autosomal recessive otofaciocervical syndrome type 2 with novel homozygous small insertion in PAX1 gene. Am. J. Med. Genet. A. 2018;176:1200–1206. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

