β-Elemene induced ferroptosis via TFEB-mediated GPX4 degradation in EGFR wide-type non-small cell lung cancer
- PMID: 37689240
- PMCID: PMC11331178
- DOI: 10.1016/j.jare.2023.08.018
β-Elemene induced ferroptosis via TFEB-mediated GPX4 degradation in EGFR wide-type non-small cell lung cancer
Abstract
Introduction: β-Elemene (β-ELE), derived from Curcuma wenyujin, has anticancer effect on non-small cell lung cancer (NSCLC). However, the potential target and detail mechanism were still not clear. TFEB is the master regulator of lysosome biogenesis. Ferroptosis, a promising strategy for cancer therapy could be triggered via suppression on glutathione peroxidase 4 (GPX4). Weather TFEB-mediated lysosome degradation contributes to GPX4 decline and how β-ELE modulates on this process are not clear.
Objectives: To observe the action of β-ELE on TFEB, and the role of TFEB-mediated GPX4 degradation in β-ELE induced ferroptosis.
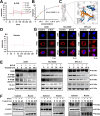
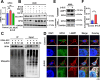
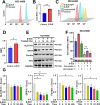
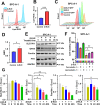
Methods: Surface plasmon resonance (SPR) and molecular docking were applied to observe the binding affinity of β-ELE on TFEB. Activation of TFEB and lysosome were observed by immunofluorescence, western blot, flow cytometry and qPCR. Ferroptosis induced by β-ELE was observed via lipid ROS, a labile iron pool (LIP) assay and western blot. A549TFEB KO cells were established via CRISPR/Cas9. The regulation of TFEB on GPX4 and ferroptosis was observed in β-ELE treated A549WT and A549TFEB KO cells, which was further studied in orthotopic NOD/SCID mouse model.
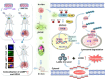
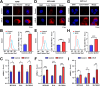
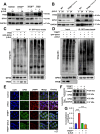
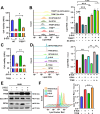
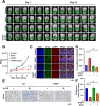
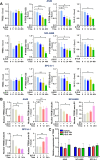
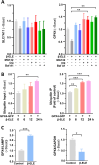
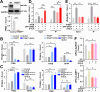
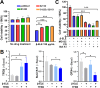
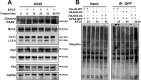
Results: β-ELE can bind to TFEB, notably activate TFEB, lysosome and transcriptional increase on downstream gene GLA, MCOLN1, SLC26A11 involved in lysosome activity in EGFR wild-type NSCLC cells. β-ELE increased GPX4 ubiquitination and lysosomal localization, with the increase on lysosome degradation of GPX4. Furthermore, β-ELE induced ferroptosis, which could be promoted by TFEB overexpression or compromised by TFEB knockout. Genetic knockout or inactivation of TFEB compromised β-ELE induced lysosome degradation of GPX4, which was further demonstrated in orthotopic NSCLC NOD/SCID mice model.
Conclusion: This study firstly demonstrated that TFEB promoted GPX4 lysosome degradation contributes to β-ELE induced ferroptosis in EGFR wild-type NSCLC, which gives a clue that TFEB mediated GPX4 degradation would be a novel strategy for ferroptosis induction and NSCLC therapy.
Keywords: Ferroptosis; Lysosome; Non-small cell lung cancer; TFEB; β-Elemene.
Copyright © 2024. Production and hosting by Elsevier B.V.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

