FBXO21 mediated degradation of p85α regulates proliferation and survival of acute myeloid leukemia
- PMID: 37689825
- PMCID: PMC10624613
- DOI: 10.1038/s41375-023-02020-w
FBXO21 mediated degradation of p85α regulates proliferation and survival of acute myeloid leukemia
Abstract
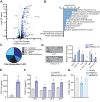
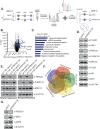
Acute myeloid leukemia (AML) is a heterogeneous disease characterized by clonal expansion of myeloid blasts in the bone marrow (BM). Despite advances in therapy, the prognosis for AML patients remains poor, and there is a need to identify novel molecular pathways regulating tumor cell survival and proliferation. F-box ubiquitin E3 ligase, FBXO21, has low expression in AML, but expression correlates with survival in AML patients and patients with higher expression have poorer outcomes. Silencing FBXO21 in human-derived AML cell lines and primary patient samples leads to differentiation, inhibition of tumor progression, and sensitization to chemotherapy agents. Additionally, knockdown of FBXO21 leads to up-regulation of cytokine signaling pathways. Through a mass spectrometry-based proteomic analysis of FBXO21 in AML, we identified that FBXO21 ubiquitylates p85α, a regulatory subunit of the phosphoinositide 3-kinase (PI3K) pathway, for degradation resulting in decreased PI3K signaling, dimerization of free p85α and ERK activation. These findings reveal the ubiquitin E3 ligase, FBXO21, plays a critical role in regulating AML pathogenesis, specifically through alterations in PI3K via regulation of p85α protein stability.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

