A predictive model of immune infiltration and prognosis of head and neck squamous cell carcinoma based on cell adhesion-related genes: including molecular biological validation
- PMID: 37691922
- PMCID: PMC10484396
- DOI: 10.3389/fimmu.2023.1190678
A predictive model of immune infiltration and prognosis of head and neck squamous cell carcinoma based on cell adhesion-related genes: including molecular biological validation
Abstract
Background: Focal adhesion serves as a bridge between tumour cells and the extracellular matrix (ECM) and has multiple roles in tumour invasion, migration, and therapeutic resistance. However, studies on focal adhesion-related genes (FARGs) in head and neck squamous cell carcinoma (HNSCC) are limited.
Methods: Data on HNSCC samples were obtained from The Cancer Genome Atlas and GSE41613 datasets, and 199 FARGs were obtained from the Molecular Signatures database. The integrated datasets' dimensions were reduced by the use of cluster analysis, which was also used to classify patients with HNSCC into subclusters. A FARG signature model was developed and utilized to calculate each patient's risk score using least extreme shrinkage and selection operator regression analysis. The risk score was done to quantify the subgroups of all patients. We evaluated the model's value for prognostic prediction, immune infiltration status, and therapeutic response in HNSCC. Preliminary molecular and biological experiments were performed to verify these results.
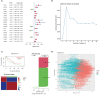
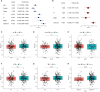
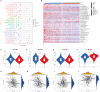
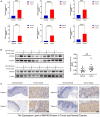
Results: Two different HNSCC molecular subtypes were identified according to FARGs, and patients with C2 had a shorter overall survival (OS) than those with C1. We constructed an FARG signature comprising nine genes. We constructed a FARG signature consisting of nine genes. Patients with higher risk scores calculated from the FARG signature had a lower OS, and the FARG signature was considered an independent prognostic factor for HNSCC in univariate and multivariate analyses. FARGs are associated with immune cell invasion, gene mutation status, and chemosensitivity. Finally, we observed an abnormal overexpression of MAPK9 in HNSCC tissues, and MAPK9 knockdown greatly impeded the proliferation, migration, and invasion of HNSCC cells.
Conclusion: The FARG signature can provide reliable prognostic prediction for patients with HNSCC. Apart from that, the genes in this model were related to immune invasion, gene mutation status, and chemosensitivity, which may provide new ideas for targeted therapies for HNSCC.
Keywords: chemotherapy sensitivity; focal adhesion-related gene; head and neck squamous cell carcinoma; immune infiltration; prognosis.
Copyright © 2023 Liu, Wu, Fu, Han, Wang, Zhang, Liang, Tao, Zhang, Shen, Xu, Yin, Chen, Liu, Pan, Liang and Wu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures









Similar articles
-
Transcriptome analysis reveals the prognostic and immune infiltration characteristics of glycolysis and hypoxia in head and neck squamous cell carcinoma.BMC Cancer. 2022 Mar 31;22(1):352. doi: 10.1186/s12885-022-09449-9. BMC Cancer. 2022. PMID: 35361159 Free PMC article.
-
Inflammatory response-related genes predict prognosis in patients with HNSCC.Immunol Lett. 2023 Jul;259:46-60. doi: 10.1016/j.imlet.2023.06.003. Epub 2023 Jun 4. Immunol Lett. 2023. PMID: 37279805
-
Comprehensive FGFR3 alteration-related transcriptomic characterization is involved in immune infiltration and correlated with prognosis and immunotherapy response of bladder cancer.Front Immunol. 2022 Jul 26;13:931906. doi: 10.3389/fimmu.2022.931906. eCollection 2022. Front Immunol. 2022. PMID: 35958598 Free PMC article.
-
Development and experimental verification of a prognosis model for disulfidptosis-associated genes in HNSCC.Medicine (Baltimore). 2024 Mar 22;103(12):e37308. doi: 10.1097/MD.0000000000037308. Medicine (Baltimore). 2024. PMID: 38518012 Free PMC article. Review.
-
Role of Focal Adhesion Kinase in Head and Neck Squamous Cell Carcinoma and Its Therapeutic Prospect.Onco Targets Ther. 2020 Oct 9;13:10207-10220. doi: 10.2147/OTT.S270342. eCollection 2020. Onco Targets Ther. 2020. PMID: 33116602 Free PMC article. Review.
Cited by
-
Establishment and validation of a novel risk model based on PANoptosis-related genes to predict prognosis in head and neck squamous cell carcinoma.Medicine (Baltimore). 2025 May 2;104(18):e42299. doi: 10.1097/MD.0000000000042299. Medicine (Baltimore). 2025. PMID: 40324259 Free PMC article.
-
An angiogenesis-associated gene-based signature predicting prognosis and immunotherapy efficacy of head and neck squamous cell carcinoma patients.J Cancer Res Clin Oncol. 2024 Feb 12;150(2):91. doi: 10.1007/s00432-024-05606-8. J Cancer Res Clin Oncol. 2024. PMID: 38347320 Free PMC article.
-
A prognostic model for Schistosoma japonicum infection-associated liver hepatocellular carcinoma: strengthening the connection through initial biological experiments.Infect Agent Cancer. 2024 Mar 21;19(1):10. doi: 10.1186/s13027-024-00569-4. Infect Agent Cancer. 2024. PMID: 38515119 Free PMC article.
-
Construction of a prognostic signature based on T-helper 17 cells differentiation-related genes for predicting survival and tumor microenvironment in head and neck squamous cell carcinoma.Medicine (Baltimore). 2025 Jan 24;104(4):e41273. doi: 10.1097/MD.0000000000041273. Medicine (Baltimore). 2025. PMID: 39854737 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

