mRNA splicing is modulated by intronic microRNAs
- PMID: 37692287
- PMCID: PMC10492213
- DOI: 10.1016/j.isci.2023.107723
mRNA splicing is modulated by intronic microRNAs
Abstract
Splicing of transcripts is catalyzed by the spliceosome, a mega-complex consisting of hundreds of proteins and five snRNAs, which employs direct interactions. When U1 snRNA forms high-affinity binding, namely more than eight base pairs, with the 5'SS, the result is usually a suppressing effect on the splicing activity. This likely occurs due to the inefficient unwinding of U1/5'SS base-pairing or other regulatory obstructions. Here, we show in vitro and in patient-derived cell lines that pre-microRNAs can modulate the splicing reaction by interacting with U1 snRNA. This leads to reduced binding affinity to the 5'SS, and hence promotes the inclusion of exons containing 5'SS, despite sequence-based high affinity to U1. Application of the mechanism resulted in correction of the splicing defect in the disease-causing VCAN gene from an individual with Wagner syndrome. This pre-miRNA/U1 interaction can regulate the expression of alternatively spliced exons, thus extending the scope of mechanisms regulating splicing.
Keywords: Cell biology; Functional aspects of cell biology; Molecular biology.
© 2023.
Conflict of interest statement
The authors declare no competing interests.
Figures

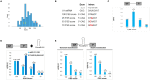
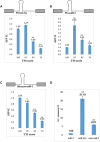

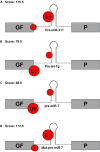
References
-
- Deckert J., Hartmuth K., Boehringer D., Behzadnia N., Will C.L., Kastner B., Stark H., Urlaub H., Lührmann R. Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol. Cell Biol. 2006;26:5528–5543. doi: 10.1128/MCB.00582-06. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

