The core bacteriobiome of Côte d'Ivoire soils across three vegetation zones
- PMID: 37692382
- PMCID: PMC10483230
- DOI: 10.3389/fmicb.2023.1220655
The core bacteriobiome of Côte d'Ivoire soils across three vegetation zones
Abstract
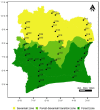
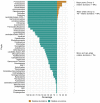
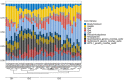
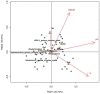
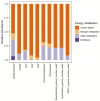
The growing understanding that soil bacteria play a critical role in ecosystem servicing has led to a number of large-scale biogeographical surveys of soil microbial diversity. However, most of such studies have focused on northern hemisphere regions and little is known of either the detailed structure or function of soil microbiomes of sub-Saharan African countries. In this paper, we report the use of high-throughput amplicon sequencing analyses to investigate the biogeography of soil bacteria in soils of Côte d'Ivoire. 45 surface soil samples were collected from Côte d'Ivoire, representing all major biomes, and bacterial community composition was assessed by targeting the V4-V5 hypervariable region of the 16S ribosomal RNA gene. Causative relationships of both soil physicochemical properties and climatic data on bacterial community structure were infered. 48 phyla, 92 classes, 152 orders, 356 families, and 1,234 genera of bacteria were identified. The core bacteriobiome consisted of 10 genera ranked in the following order of total abundance: Gp6, Gaiella, Spartobacteria_genera_incertae_sedis, WPS-1_genera_incertae_sedis, Gp4, Rhodoplanes, Pseudorhodoplanes, Bradyrhizobium, Subdivision3_genera_incertae_sedis, and Gp3. Some of these genera, including Gp4 and WPS-1_genera_incertae_sedis, were unequally distributed between forest and savannah areas while other taxa (Bradyrhizobium and Rhodoplanes) were consistently found in all biomes. The distribution of the core genera, together with the 10 major phyla, was influenced by several environmental factors, including latitude, pH, Al and K. The main pattern of distribution that was observed for the core bacteriobiome was the vegetation-independent distribution scheme. In terms of predicted functions, all core bacterial taxa were involved in assimilatory sulfate reduction, while atmospheric dinitrogen (N2) reduction was only associated with the genus Bradyrhizobium. This work, which is one of the first such study to be undertaken at this scale in Côte d'Ivoire, provides insights into the distribution of bacterial taxa in Côte d'Ivoire soils, and the findings may serve as biological indicator for land management in Côte d'Ivoire.
Keywords: 16S rDNA; Côte d’Ivoire; bacteria; common core bacteriobiome; microbiome.
Copyright © 2023 Amon, Fossou, Ebou, Koua, Kouadjo, Brou, Voko Bi, Cowan and Zézé.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Albuquerque L., França L., Rainey F. A., Schumann P., Nobre M. F., da Costa M. S. (2011). Gaiella occulta gen. Nov., sp. nov., a novel representative of a deep branching phylogenetic lineage within the class Actinobacteria and proposal of Gaiellaceae fam. Nov. and Gaiellales Ord. Nov. Syst. Appl. Microbiol. 34, 595–599. doi: 10.1016/j.syapm.2011.07.001, PMID: - DOI - PubMed
-
- Avenard J.-M., Eldin M., Girard G., Sircoulon J., Touchebeuf P., Guillaumet J.-L., et al. (1971). Le milieu naturel de la Côte d’Ivoire, Paris, ORSTOM
Publication types
LinkOut - more resources
Full Text Sources

