Single-cell RNA sequencing in juvenile idiopathic arthritis
- PMID: 37692495
- PMCID: PMC10491939
- DOI: 10.1016/j.gendis.2023.04.014
Single-cell RNA sequencing in juvenile idiopathic arthritis
Abstract
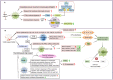
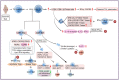
Juvenile idiopathic arthritis (JIA) is one of the most common chronic inflammatory rheumatic diseases in children, with onset before age 16 and lasting for more than 6 weeks. JIA is a highly heterogeneous condition with various consequences for health and quality of life. For some JIA patients, early detection and intervention remain challenging. As a result, further investigation of the complex and unknown mechanisms underlying JIA is required. Advances in technology now allow us to describe the biological heterogeneity and function of individual cell populations in JIA. Through this review, we hope to provide novel ideas and potential targets for the diagnosis and treatment of JIA by summarizing the current findings of single-cell RNA sequencing studies and understanding how the major cell subsets drive JIA pathogenesis.
Keywords: Juvenile idiopathic arthritis; Macrophage; Macrophage activation syndrome; Monocyte; Single-cell RNA sequencing; Synoviocyte; T cell.
© 2023 The Authors. Publishing services by Elsevier B.V. on behalf of KeAi Communications Co., Ltd.
Figures

Similar articles
-
B Cells on the Stage of Inflammation in Juvenile Idiopathic Arthritis: Leading or Supporting Actors in Disease Pathogenesis?Front Med (Lausanne). 2022 Apr 4;9:851532. doi: 10.3389/fmed.2022.851532. eCollection 2022. Front Med (Lausanne). 2022. PMID: 35449805 Free PMC article. Review.
-
Increased Development of Th1, Th17, and Th1.17 Cells Under T1 Polarizing Conditions in Juvenile Idiopathic Arthritis.Front Immunol. 2022 Jul 4;13:848168. doi: 10.3389/fimmu.2022.848168. eCollection 2022. Front Immunol. 2022. PMID: 35860254 Free PMC article.
-
Juvenile idiopathic arthritis: Diagnosis and differential diagnosis.Korean J Pediatr. 2010 Nov;53(11):931-5. doi: 10.3345/kjp.2010.53.11.931. Epub 2010 Nov 30. Korean J Pediatr. 2010. PMID: 21218014 Free PMC article.
-
Differential Accumulation and Activation of Monocyte and Dendritic Cell Subsets in Inflamed Synovial Fluid Discriminates Between Juvenile Idiopathic Arthritis and Septic Arthritis.Front Immunol. 2020 Jul 31;11:1716. doi: 10.3389/fimmu.2020.01716. eCollection 2020. Front Immunol. 2020. PMID: 32849606 Free PMC article.
-
New advances in juvenile idiopathic arthritis.Chang Gung Med J. 2012 Jan-Feb;35(1):1-14. doi: 10.4103/2319-4170.106171. Chang Gung Med J. 2012. PMID: 22483423 Review.
Cited by
-
Potential marker genes for chronic obstructive pulmonary disease revealed based on single-cell sequencing and Mendelian randomization analysis.Aging (Albany NY). 2024 May 23;16(10):8922-8943. doi: 10.18632/aging.205849. Epub 2024 May 23. Aging (Albany NY). 2024. PMID: 38787375 Free PMC article.
-
The emerging role of neutrophil extracellular traps in autoimmune and autoinflammatory diseases.MedComm (2020). 2025 Mar 6;6(3):e70101. doi: 10.1002/mco2.70101. eCollection 2025 Mar. MedComm (2020). 2025. PMID: 40060194 Free PMC article. Review.
-
Comprehensive analysis of juvenile idiopathic arthritis patients' immune characteristics based on bulk and single-cell sequencing data.Front Mol Biosci. 2024 May 1;11:1359235. doi: 10.3389/fmolb.2024.1359235. eCollection 2024. Front Mol Biosci. 2024. PMID: 38751447 Free PMC article.
-
NLRP3 overexpression exacerbated synovium tissue degeneration in juvenile collagen-induced arthritis.Sci Rep. 2025 Feb 27;15(1):7024. doi: 10.1038/s41598-025-86720-6. Sci Rep. 2025. PMID: 40016261 Free PMC article.
References
-
- Thierry S., Fautrel B., Lemelle I., Guillemin F. Prevalence and incidence of juvenile idiopathic arthritis: a systematic review. Joint Bone Spine. 2014;81(2):112–117. - PubMed
-
- Petty R.E., Southwood T.R., Manners P., et al. International League of Associations for Rheumatology classification of juvenile idiopathic arthritis: second revision, Edmonton, 2001. J Rheumatol. 2004;31(2):390–392. - PubMed
-
- Thatayatikom A., Modica R., De Leucio A. StatPearls Publishing; Treasure Island (FL): 2023. Juvenile idiopathic arthritis. (StatPearls). - PubMed
-
- Lee J.J.Y., Schneider R. Systemic juvenile idiopathic arthritis. Pediatr Clin. 2018;65(4):691–709. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials

