From vaccine to pathogen: Modeling Sabin 2 vaccine virus reversion and evolutionary epidemiology in Matlab, Bangladesh
- PMID: 37692896
- PMCID: PMC10491863
- DOI: 10.1093/ve/vead044
From vaccine to pathogen: Modeling Sabin 2 vaccine virus reversion and evolutionary epidemiology in Matlab, Bangladesh
Abstract
The oral poliovirus vaccines (OPVs) are one of the most effective disease eradication tools in public health. However, the OPV strains are genetically unstable and can cause outbreaks of circulating, vaccine-derived Type 2 poliovirus (cVDPV2) that are clinically indistinguishable from wild poliovirus (WPV) outbreaks. Here, we developed a Sabin 2 reversion model that simulates the reversion of Sabin 2 to reacquire a WPV-like phenotype based on the clinical differences in shedding duration and infectiousness between individuals vaccinated with Sabin 2 and those infected with WPV. Genetic reversion is informed by a canonical reversion pathway defined by three gatekeeper mutations (A481G, U2909C, and U398C) and the accumulation of deleterious nonsynonymous mutations. Our model captures essential aspects of both phenotypic and molecular evolution and simulates transmission using a multiscale transmission model that consolidates the relationships among immunity, susceptibility, and transmission risk. Despite rapid Sabin 2 attenuation reversal, we show that the emergence of a revertant virus does not guarantee a cVDPV2 outbreak. When simulating outbreaks in Matlab, Bangladesh, we found that cVDPV2 outbreaks are most likely in areas with low population-level immunity and poor sanitation. In Matlab, our model predicted that declining immunity against Type 2 poliovirus following the cessation of routine OPV vaccination was not enough to promote cVDPV2 emergence. However, cVDPV2 emergencedepended on the average viral exposure dose per contact, which was modeled as a combination of the viral concentration per fecal gram and the average fecal-oral dose per contact. These results suggest that cVDPV2 emergence risk can be mitigated by reducing the amount of infectious fecal material individuals are exposed to. Thus, a combined strategy of assessing and improving sanitation levels in conjunction with high-coverage vaccination campaigns could limit the future cVDPV2 emergence.
Keywords: Sabin 2; cVDPV2; evolution; genetic reversion; modeling; poliovirus; vaccine.
© The Author(s) 2023. Published by Oxford University Press.
Conflict of interest statement
The authors declare no competing interests during the study. W.W., J.G., and M.F. were employed by the funders through the Bill and Melinda Gates Foundation after the study completion.
Figures


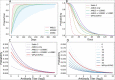
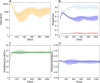
References
-
- Abraham R. et al. (1993) ‘Shedding of Virulent Poliovirus Revertants during Immunization with Oral Poliovirus Vaccine after Prior Immunization with Inactivated Polio Vaccine’, Journal of Infectious Diseases, 168: 1105–9. - PubMed
-
- Almond J. W. (1987) ‘The Attenuation of Poliovirus Neurovirulence’, Annual Review of Microbiology, 41: 153–80. - PubMed
-
- Bandyopadhyay A. S. et al. (2015) ‘Polio Vaccination: Past, Present and Future’, Future Microbiology, 10: 791–808. - PubMed
LinkOut - more resources
Full Text Sources

