This is a preprint.
LINC01638 Sustains Human Mesenchymal Stem Cell Self-Renewal and Competency for Osteogenic Cell Fate
- PMID: 37693373
- PMCID: PMC10491330
- DOI: 10.21203/rs.3.rs-3210911/v1
LINC01638 Sustains Human Mesenchymal Stem Cell Self-Renewal and Competency for Osteogenic Cell Fate
Update in
-
LINC01638 sustains human mesenchymal stem cell self-renewal and competency for osteogenic cell fate.Sci Rep. 2023 Nov 20;13(1):20314. doi: 10.1038/s41598-023-46202-z. Sci Rep. 2023. PMID: 37985890 Free PMC article.
Abstract
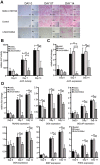
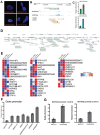
The skeleton forms from multipotent human mesenchymal stem cells (hMSCs) competent to commit to specific lineages. Long noncoding RNAs (lncRNAs) have been identified as key epigenetic regulators of tissue development. However, regulation of osteogenesis by lncRNAs as mediators of commitment to the bone phenotype is largely unexplored. We focused on LINC01638, which is highly expressed in hMSCs and has been studied in cancers, but not in regulating osteogenesis. We demonstrated that LINC01638 promotes initiation of the osteoblast phenotype. Our findings reveal that LINC01638 is present at low levels during the induction of osteoblast differentiation. CRISPRi knockdown of LINC01638 in MSCs prevents osteogenesis and alkaline phosphatase expression, inhibiting osteoblast differentiation. This resulted in decreased MSC cell growth rate, accompanied by double-strand breaks, DNA damage, and cell senescence. Transcriptome profiling of control and LINC01638-depleted hMSCs identified > 2,000 differentially expressed mRNAs related to cell cycle, cell division, spindle formation, DNA repair, and osteogenesis. Using ChIRP-qPCR, molecular mechanisms of chromatin interactions revealed the LINC01638 locus (Chr 22) includes many lncRNAs and bone-related genes. These novel findings identify the obligatory role for LINC01638 to sustain MSC pluripotency regulating osteoblast commitment and growth, as well as for physiological remodeling of bone tissue.
Keywords: LINC01638; RNA-Seq; gene expression; long noncoding RNA; mesenchymal stromal cells; osteoblast; osteogenesis; proliferation; senescence.
Figures



References
-
- Hemming S, Cakouros D, Isenmann S, Cooper L, Menicanin D, Zannettino A, Gronthos S: EZH2 and KDM6A act as an epigenetic switch to regulate mesenchymal stem cell lineage specification. Stem Cells 2014, 32:802–815. - PubMed
-
- Guo Q, Guo Q, Xiao Y, Li C, Huang Y, Luo X: Regulation of bone marrow mesenchymal stem cell fate by long non-coding RNA. Bone 2020, 141:115617. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

