This is a preprint.
Tick hemocytes have pleiotropic roles in microbial infection and arthropod fitness
- PMID: 37693411
- PMCID: PMC10491215
- DOI: 10.1101/2023.08.31.555785
Tick hemocytes have pleiotropic roles in microbial infection and arthropod fitness
Update in
-
Tick hemocytes have a pleiotropic role in microbial infection and arthropod fitness.Nat Commun. 2024 Mar 8;15(1):2117. doi: 10.1038/s41467-024-46494-3. Nat Commun. 2024. PMID: 38459063 Free PMC article.
Abstract
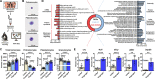
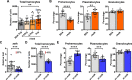
Uncovering the complexity of systems in non-model organisms is critical for understanding arthropod immunology. Prior efforts have mostly focused on Dipteran insects, which only account for a subset of existing arthropod species in nature. Here, we describe immune cells or hemocytes from the clinically relevant tick Ixodes scapularis using bulk and single cell RNA sequencing combined with depletion via clodronate liposomes, RNA interference, Clustered Regularly Interspaced Short Palindromic Repeats activation (CRISPRa) and RNA-fluorescence in situ hybridization (FISH). We observe molecular alterations in hemocytes upon tick infestation of mammals and infection with either the Lyme disease spirochete Borrelia burgdorferi or the rickettsial agent Anaplasma phagocytophilum. We predict distinct hemocyte lineages and reveal clusters exhibiting defined signatures for immunity, metabolism, and proliferation during hematophagy. Furthermore, we perform a mechanistic characterization of two I. scapularis hemocyte markers: hemocytin and astakine. Depletion of phagocytic hemocytes affects hemocytin and astakine levels, which impacts blood feeding and molting behavior of ticks. Hemocytin specifically affects the c-Jun N-terminal kinase (JNK) signaling pathway, whereas astakine alters hemocyte proliferation in I. scapularis. Altogether, we uncover the heterogeneity and pleiotropic roles of hemocytes in ticks and provide a valuable resource for comparative biology in arthropods.
Keywords: Anaplasma phagocytophilum; Borrelia burgdorferi; Hematophagy; Hemocytes; Tick-Borne Diseases.
Figures






References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
